The kinetic mechanisms of fast-decay red-fluorescent genetically encoded calcium indicators
- PMID: 30651353
- PMCID: PMC6422079
- DOI: 10.1074/jbc.RA118.004543
The kinetic mechanisms of fast-decay red-fluorescent genetically encoded calcium indicators
Abstract
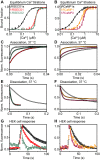
Genetically encoded calcium indicators (GECIs) are useful reporters of cell-signaling, neuronal, and network activities. We have generated novel fast variants and investigated the kinetic mechanisms of two recently developed red-fluorescent GECIs (RGECIs), mApple-based jRGECO1a and mRuby-based jRCaMP1a. In the formation of fluorescent jRGECO1a and jRCaMP1a complexes, calcium binding is followed by rate-limiting isomerization. However, fluorescence decay of calcium-bound jRGECO1a follows a different pathway from its formation: dissociation of calcium occurs first, followed by the peptide, similarly to GCaMP-s. In contrast, fluorescence decay of calcium-bound jRCaMP1a occurs by the reversal of the on-pathway: peptide dissociation is followed by calcium. The mechanistic differences explain the generally slower off-kinetics of jRCaMP1a-type indicators compared with GCaMP-s and jRGECO1a-type GECI: the fluorescence decay rate of f-RCaMP1 was 21 s-1, compared with 109 s-1 for f-RGECO1 and f-RGECO2 (37 °C). Thus, the CaM-peptide interface is an important determinant of the kinetic responses of GECIs; however, the topology of the structural link to the fluorescent protein demonstrably affects the internal dynamics of the CaM-peptide complex. In the dendrites of hippocampal CA3 neurons, f-RGECO1 indicates calcium elevation in response to a 100 action potential train in a linear fashion, making the probe particularly useful for monitoring large-amplitude, fast signals, e.g. those in dendrites, muscle cells, and immune cells.
Keywords: biosensor; calcium; calcium imaging; fluorescence; kinetics.
© 2019 Kerruth et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

