Growth suppression by dual BRAF(V600E) and NRAS(Q61) oncogene expression is mediated by SPRY4 in melanoma
- PMID: 30651601
- PMCID: PMC6756020
- DOI: 10.1038/s41388-018-0632-2
Growth suppression by dual BRAF(V600E) and NRAS(Q61) oncogene expression is mediated by SPRY4 in melanoma
Abstract
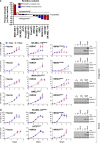
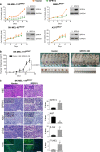
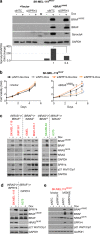
The underlying forces that shape mutational patterns within any type of cancer have been poorly characterized. One of the best preserved exclusionary relationships is that between BRAF(V600E) and NRAS(Q61) in melanomas. To explore possible mechanisms which could explain this phenomenon, we overexpressed NRAS(Q61) in a set of BRAF(V600E) melanoma lines and vice versa. Controlled expression of a second activating oncogene led to growth arrest ("synthetic suppression") in a subset of cells, which was accompanied by cell cycle arrest and senescence in several melanoma cell lines along with apoptosis. Through differential gene expression analysis, we identified SPRY4 as the potential mediator of this synthetic response to dual oncogene suppression. Ectopic introduction of SPRY4 recapitulated the growth arrest phenotype of dual BRAF(V600E)/NRAS(Q61) expression while SPRY4 depletion led to a partial rescue from oncogenic antagonism. This study thus defined SPRY4 as a potential mediator of synthetic suppression, which is likely to contribute to the observed exclusivity between BRAF(V600E) and NRAS(Q61R) mutations in melanoma. Further leverage of the SPRY4 pathway may also hold therapeutic promise for NRAS(Q61) melanomas.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Vemurafenib Drives Epithelial-to-Mesenchymal Transition Gene Expression in BRAF Inhibitor‒Resistant BRAFV600E/NRASQ61K Melanoma Enhancing Tumor Growth and Metastasis in a Bioluminescent Murine Model.J Invest Dermatol. 2022 May;142(5):1456-1465.e1. doi: 10.1016/j.jid.2021.10.007. Epub 2021 Oct 21. J Invest Dermatol. 2022. PMID: 34687745 Free PMC article.
-
Mefloquine induces ER stress and apoptosis in BRAFi-resistant A375-BRAFV600E /NRASQ61K malignant melanoma cells targeting intracranial tumors in a bioluminescent murine model.Mol Carcinog. 2022 Jun;61(6):603-614. doi: 10.1002/mc.23407. Epub 2022 Apr 13. Mol Carcinog. 2022. PMID: 35417045 Free PMC article.
-
BRAF(V600E) and NRAS(Q61L/Q61R) mutation analysis in metastatic melanoma using immunohistochemistry: a study of 754 cases highlighting potential pitfalls and guidelines for interpretation and reporting.Histopathology. 2016 Oct;69(4):680-6. doi: 10.1111/his.12992. Epub 2016 Jun 15. Histopathology. 2016. PMID: 27151331
-
NRAS mutant melanoma: biological behavior and future strategies for therapeutic management.Oncogene. 2013 Jun 20;32(25):3009-18. doi: 10.1038/onc.2012.453. Epub 2012 Oct 15. Oncogene. 2013. PMID: 23069660 Free PMC article. Review.
-
Germline MC1R variants and frequency of somatic BRAF, NRAS, and TERT mutations in melanoma: Literature review and meta-analysis.Mol Carcinog. 2021 Mar;60(3):167-171. doi: 10.1002/mc.23280. Epub 2021 Jan 14. Mol Carcinog. 2021. PMID: 33444485 Review.
Cited by
-
Vemurafenib Drives Epithelial-to-Mesenchymal Transition Gene Expression in BRAF Inhibitor‒Resistant BRAFV600E/NRASQ61K Melanoma Enhancing Tumor Growth and Metastasis in a Bioluminescent Murine Model.J Invest Dermatol. 2022 May;142(5):1456-1465.e1. doi: 10.1016/j.jid.2021.10.007. Epub 2021 Oct 21. J Invest Dermatol. 2022. PMID: 34687745 Free PMC article.
-
Mefloquine induces ER stress and apoptosis in BRAFi-resistant A375-BRAFV600E /NRASQ61K malignant melanoma cells targeting intracranial tumors in a bioluminescent murine model.Mol Carcinog. 2022 Jun;61(6):603-614. doi: 10.1002/mc.23407. Epub 2022 Apr 13. Mol Carcinog. 2022. PMID: 35417045 Free PMC article.
-
SPRY4 inhibits and sensitizes the primary KIT mutants in gastrointestinal stromal tumors (GISTs) to imatinib.Gastric Cancer. 2023 Sep;26(5):677-690. doi: 10.1007/s10120-023-01402-4. Epub 2023 May 24. Gastric Cancer. 2023. PMID: 37222910
-
Signaling Switching from Hedgehog-GLI to MAPK Signaling Potentially Serves as a Compensatory Mechanism in Melanoma Cell Lines Resistant to GANT-61.Biomedicines. 2023 May 3;11(5):1353. doi: 10.3390/biomedicines11051353. Biomedicines. 2023. PMID: 37239024 Free PMC article.
-
Detection of Gene Mutations in Liquid Biopsy of Melanoma Patients: Overview and Future Perspectives.Curr Treat Options Oncol. 2020 Feb 11;21(3):19. doi: 10.1007/s11864-020-0708-4. Curr Treat Options Oncol. 2020. PMID: 32048063 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

