Excised linear introns regulate growth in yeast
- PMID: 30651636
- PMCID: PMC6464110
- DOI: 10.1038/s41586-018-0828-1
Excised linear introns regulate growth in yeast
Abstract
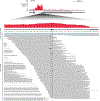
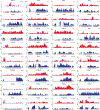
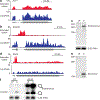
Spliceosomal introns are ubiquitous non-coding RNAs that are typically destined for rapid debranching and degradation. Here we describe 34 excised introns in Saccharomyces cerevisiae that-despite being rapidly degraded in log-phase growth-accumulate as linear RNAs under either saturated-growth conditions or other stresses that cause prolonged inhibition of TORC1, which is a key integrator of growth signalling. Introns that become stabilized remain associated with components of the spliceosome and differ from other spliceosomal introns in having a short distance between their lariat branch point and 3' splice site, which is necessary and sufficient for their stabilization. Deletion of these unusual introns is disadvantageous in saturated conditions and causes aberrantly high growth rates in yeast that are chronically challenged with the TORC1 inhibitor rapamycin. The reintroduction of native or engineered stable introns suppresses this aberrant rapamycin response. Thus, excised introns function within the TOR growth-signalling network of S. cerevisiae and, more generally, excised spliceosomal introns can have biological functions.
Conflict of interest statement
Competing Interests
The authors declare no competing financial interests.
Figures

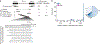




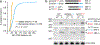
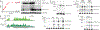

Comment in
-
Intron RNA sequences help yeast cells to survive starvation.Nature. 2019 Jan;565(7741):578-579. doi: 10.1038/d41586-019-00088-y. Nature. 2019. PMID: 30683935 Free PMC article.
-
Craving for Introns.Mol Cell. 2019 Mar 21;73(6):1095-1096. doi: 10.1016/j.molcel.2019.03.008. Mol Cell. 2019. PMID: 30901563
Similar articles
-
trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing.Mol Cell. 2007 Jun 22;26(6):883-90. doi: 10.1016/j.molcel.2007.05.020. Mol Cell. 2007. PMID: 17588521 Free PMC article.
-
Ubiquitin regulates TORC1 in yeast Saccharomyces cerevisiae.Mol Microbiol. 2016 Apr;100(2):303-14. doi: 10.1111/mmi.13319. Epub 2016 Mar 10. Mol Microbiol. 2016. PMID: 26700129 Free PMC article.
-
The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae.Genes Dev. 2017 Apr 1;31(7):702-717. doi: 10.1101/gad.295188.116. Genes Dev. 2017. PMID: 28446598 Free PMC article.
-
Group II intron lariat: Structural insights into the spliceosome.RNA Biol. 2015;12(9):913-7. doi: 10.1080/15476286.2015.1066956. RNA Biol. 2015. PMID: 26121424 Free PMC article. Review.
-
Novel Links between TORC1 and Traditional Non-Coding RNA, tRNA.Genes (Basel). 2020 Aug 19;11(9):956. doi: 10.3390/genes11090956. Genes (Basel). 2020. PMID: 32825021 Free PMC article. Review.
Cited by
-
Nutrient-dependent regulation of a stable intron modulates germline mitochondrial quality control.Nat Commun. 2024 Feb 10;15(1):1252. doi: 10.1038/s41467-024-45651-y. Nat Commun. 2024. PMID: 38341415 Free PMC article.
-
Intronic regions of the human coagulation factor VIII gene harboring transcription factor binding sites with a strong bias towards the short-interspersed elements.Heliyon. 2020 Sep 2;6(9):e04727. doi: 10.1016/j.heliyon.2020.e04727. eCollection 2020 Sep. Heliyon. 2020. PMID: 32944665 Free PMC article.
-
Identification of Alternative Polyadenylation in Cyanidioschyzon merolae Through Long-Read Sequencing of mRNA.Front Genet. 2022 Jan 28;12:818697. doi: 10.3389/fgene.2021.818697. eCollection 2021. Front Genet. 2022. PMID: 35154260 Free PMC article.
-
Transcriptional regulation of genes bearing intronic heterochromatin in the rice genome.PLoS Genet. 2020 Mar 18;16(3):e1008637. doi: 10.1371/journal.pgen.1008637. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32187179 Free PMC article.
-
Quiescence in Saccharomyces cerevisiae.Annu Rev Genet. 2022 Nov 30;56:253-278. doi: 10.1146/annurev-genet-080320-023632. Annu Rev Genet. 2022. PMID: 36449357 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

