Investigation of ( S)-(-)-Acidomycin: A Selective Antimycobacterial Natural Product That Inhibits Biotin Synthase
- PMID: 30652474
- PMCID: PMC6724193
- DOI: 10.1021/acsinfecdis.8b00345
Investigation of ( S)-(-)-Acidomycin: A Selective Antimycobacterial Natural Product That Inhibits Biotin Synthase
Abstract
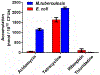
The synthesis, absolute stereochemical configuration, complete biological characterization, mechanism of action and resistance, and pharmacokinetic properties of ( S)-(-)-acidomycin are described. Acidomycin possesses promising antitubercular activity against a series of contemporary drug susceptible and drug-resistant M. tuberculosis strains (minimum inhibitory concentrations (MICs) = 0.096-6.2 μM) but is inactive against nontuberculosis mycobacteria and Gram-positive and Gram-negative pathogens (MICs > 1000 μM). Complementation studies with biotin biosynthetic pathway intermediates and subsequent biochemical studies confirmed acidomycin inhibits biotin synthesis with a Ki of approximately 1 μM through the competitive inhibition of biotin synthase (BioB) and also stimulates unproductive cleavage of S-adenosyl-l-methionine (SAM) to generate the toxic metabolite 5'-deoxyadenosine. Cell studies demonstrate acidomycin selectively accumulates in M. tuberculosis providing a mechanistic basis for the observed antibacterial activity. The development of spontaneous resistance by M. tuberculosis to acidomycin was difficult, and only low-level resistance to acidomycin was observed by overexpression of BioB. Collectively, the results provide a foundation to advance acidomycin and highlight BioB as a promising target.
Keywords: Mycobacterium tuberculosis; accumulation; acidomycin; antimetabolite; biotin biosynthesis; biotin synthase; tuberculosis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







References
-
- WHO Global tuberculosis report; World Health Organization; Geneva, Switzerland, 2018.
-
- Organization W. H., Treatment of tuberculosis. Guidelines. 4th ed.; WHO/HTM/TB/2009.420: 2010. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

