Negative allosteric modulators of cannabinoid receptor 2: protein modeling, binding site identification and molecular dynamics simulations in the presence of an orthosteric agonist
- PMID: 30652534
- PMCID: PMC7487276
- DOI: 10.1080/07391102.2019.1567384
Negative allosteric modulators of cannabinoid receptor 2: protein modeling, binding site identification and molecular dynamics simulations in the presence of an orthosteric agonist
Abstract
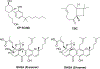
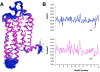
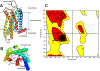
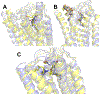
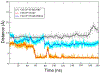
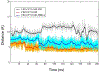
Selective activation of the cannabinoid receptor subtype 2 (CB2) shows promise for treating pain, inflammation, multiple sclerosis, cancer, ischemic/reperfusion injury and osteoporosis. Target selectivity and off-target side effects are two major limiting factors for orthosteric ligands, and therefore, the search for allosteric modulators (AMs) is a widely used drug discovery approach. To date, only a limited number of negative CB2 AMs have been identified, possessing only micromolar activity at best, and the CB2 receptor's allosteric site(s) are not well characterized. Herein, we used computational approaches including receptor modeling, site mapping, docking, molecular dynamics (MD) simulations and binding free energy calculations to predict, characterize and validate allosteric sites within the complex of the CB2 receptor with bound orthosteric agonist CP55,940. After docking of known negative CB2 allosteric modulators (NAMs), dihydro-gambogic acid (DHGA) and trans-β-caryophyllene (TBC) (note that TBC also shows agonist activity), at the predicted allosteric sites, the best total complex with CB2, CP55,940 and NAM was embedded into a hydrated lipid bilayer and subjected to a 200 ns MD simulation. The presence of an AM affected the CB2-CP55,940 complex, altering the relative positioning of the toggle switch residues and promoting a strong π-π interaction between Phe1173.36 and Trp2586.48. Binding of either TBC or DHGA to a putative allosteric pocket directly adjacent to the orthosteric ligand reduced the binding free energy of CP55,940, which is consistent with the expected effect of a negative AM. The identified allosteric sites present immense scope for the discovery of novel classes of CB2 AMs.
Keywords: CB2 allosteric modulator; CB2 receptor; G-protein-coupled receptor; docking; molecular dynamics.
Figures

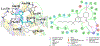
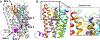
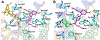
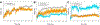


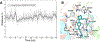
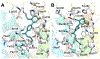
References
-
- Abood ME (2016). Allosteric Modulators: A Side Door. J Med Chem, 59(1), pp. 42–43. doi:10.1021/acs.jmedchem.5b01824 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/26645411 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/26645411 - DOI - PubMed
-
- Alaverdashvili M, & Laprairie RB (2018). The future of type 1 cannabinoid receptor allosteric ligands. [Review]. Drug Metabolism Reviews, 50(1), pp. 14–25. doi:10.1080/03602532.2018.1428341 Retrieved from https://www.scopus.com/inward/record.uri?eid=2-s2.0-85043599300&doi=10.1... Retrieved from https://www.scopus.com/inward/record.uri?eid=2-s2.0-85043599300&doi=... - DOI - PubMed
-
- Alqarni M, Myint KZ, Tong Q, Yang P, Bartlow P, Wang L, … Xie XQ (2014). Examining the critical roles of human CB2 receptor residues Valine 3.32 (113) and Leucine 5.41 (192) in ligand recognition and downstream signaling activities. Biochem Biophys Res Commun, 452(3), pp. 334–339. doi:10.1016/j.bbrc.2014.08.048 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/25148941 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/25148941 - DOI - PMC - PubMed
-
- Bernardes WA, Lucarini R, Tozatti MG, Flauzino LG, Souza MG, Turatti IC, … Cunha WR (2010). Antibacterial activity of the essential oil from Rosmarinus officinalis and its major components against oral pathogens. Z Naturforsch C, 65(9–10), pp. 588–593. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/21138060 - PubMed
-
- Cascao R, Vidal B, Raquel H, Neves-Costa A, Figueiredo N, Gupta V, … Moita LF (2014). Potent anti-inflammatory and antiproliferative effects of gambogic acid in a rat model of antigen-induced arthritis. Mediators Inflamm, 2014, p 195327. doi:10.1155/2014/195327 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/24623960 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/24623960 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
