American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: From Inception to First Data Release and Beyond-Lessons Learned and Member Institutions' Perspectives
- PMID: 30652542
- PMCID: PMC6873906
- DOI: 10.1200/CCI.17.00083
American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: From Inception to First Data Release and Beyond-Lessons Learned and Member Institutions' Perspectives
Abstract
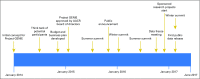
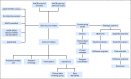
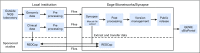
The American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) is an international data-sharing consortium focused on enabling advances in precision oncology through the gathering and sharing of tumor genetic sequencing data linked with clinical data. The project's history, operational structure, lessons learned, and institutional perspectives on participation in the data-sharing consortium are reviewed. Individuals involved with the inception and execution of AACR Project GENIE from each member institution described their experiences and lessons learned. The consortium was conceived in January 2014 and publicly released its first data set in January 2017, which consisted of 18,804 samples from 18,324 patients contributed by the eight founding institutions. Commitment and contributions from many individuals at AACR and the member institutions were crucial to the consortium's success. These individuals filled leadership, project management, informatics, data curation, contracts, ethics, and security roles. Many lessons were learned during the first 3 years of the consortium, including on how to gather, harmonize, and share data; how to make decisions and foster collaboration; and how to set the stage for continued participation and expansion of the consortium. We hope that the lessons shared here will assist new GENIE members as well as others who embark on the journey of forming a genomic data-sharing consortium.
Conflict of interest statement
Christine M. Micheel
Shawn M. Sweeney
No relationship to disclose
Michele L. LeNoue-Newton
Fabrice André
Philippe L. Bedard
Justin Guinney
Gerrit A. Meijer
Barrett J. Rollins
No relationship to disclose
Charles L. Sawyers
Nikolaus Schultz
No relationship to disclose
Kenna R. Mills Shaw
No relationship to disclose
Victor E. Velculescu
Mia A. Levy
Figures

Similar articles
-
AACR Project GENIE: Powering Precision Medicine through an International Consortium.Cancer Discov. 2017 Aug;7(8):818-831. doi: 10.1158/2159-8290.CD-17-0151. Epub 2017 Jun 1. Cancer Discov. 2017. PMID: 28572459 Free PMC article.
-
Huge Data-Sharing Project Launched.Cancer Discov. 2016 Jan;6(1):4-5. doi: 10.1158/2159-8290.CD-NB2015-159. Epub 2015 Nov 6. Cancer Discov. 2016. PMID: 26546297
-
A novel cross-disciplinary multi-institute approach to translational cancer research: lessons learned from Pennsylvania Cancer Alliance Bioinformatics Consortium (PCABC).Cancer Inform. 2007 Jun 8;3:255-74. Cancer Inform. 2007. PMID: 19455246 Free PMC article.
-
Art and Challenges of Precision Medicine: Interpreting and Integrating Genomic Data Into Clinical Practice.Am Soc Clin Oncol Educ Book. 2018 May 23;38:546-553. doi: 10.1200/EDBK_200759. Am Soc Clin Oncol Educ Book. 2018. PMID: 30231369 Review.
-
Using biointelligence to search the cancer genome: an epistemological perspective on knowledge recovery strategies to enable precision medical genomics.Oncogene. 2008 Dec;27 Suppl 2:S58-66. doi: 10.1038/onc.2009.354. Oncogene. 2008. PMID: 19956181 Review.
Cited by
-
Case Studies for Overcoming Challenges in Using Big Data in Cancer.Cancer Res. 2023 Apr 14;83(8):1183-1190. doi: 10.1158/0008-5472.CAN-22-1277. Cancer Res. 2023. PMID: 36625851 Free PMC article. Review.
-
Pan-cancer identification of clinically relevant genomic subtypes using outcome-weighted integrative clustering.Genome Med. 2020 Dec 3;12(1):110. doi: 10.1186/s13073-020-00804-8. Genome Med. 2020. PMID: 33272320 Free PMC article.
-
Key Members of the CmPn as Biomarkers Distinguish Histological and Immune Subtypes of Hepatic Cancers.Diagnostics (Basel). 2023 Mar 7;13(6):1012. doi: 10.3390/diagnostics13061012. Diagnostics (Basel). 2023. PMID: 36980321 Free PMC article.
-
Genomic alterations and possible druggable mutations in carcinoma of unknown primary (CUP).Sci Rep. 2021 Jul 23;11(1):15112. doi: 10.1038/s41598-021-94678-4. Sci Rep. 2021. PMID: 34302033 Free PMC article.
-
A network medicine approach for identifying diagnostic and prognostic biomarkers and exploring drug repurposing in human cancer.Comput Struct Biotechnol J. 2022 Nov 29;21:34-45. doi: 10.1016/j.csbj.2022.11.037. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36514340 Free PMC article.
References
-
- The AACR Project GENIE Consortium : AACR GENIE Data Guide, 2017. http://www.aacr.org/Research/Research/Documents/GENIE%20Data%20Guide.pdf
-
- The AACR Project GENIE Consortium : AACR Project GENIE Participation Evaluation Criteria, 2017. http://www.aacr.org/Documents/GENIE_New_Participant_Criteria.pdf
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

