Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry
- PMID: 30653506
- PMCID: PMC6353231
- DOI: 10.1371/journal.pgen.1007808
Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry
Abstract
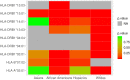
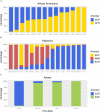
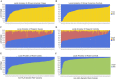
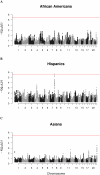
Multiple sclerosis (MS) is an autoimmune disease with high prevalence among populations of northern European ancestry. Past studies have shown that exposure to ultraviolet radiation could explain the difference in MS prevalence across the globe. In this study, we investigate whether the difference in MS prevalence could be explained by European genetic risk factors. We characterized the ancestry of MS-associated alleles using RFMix, a conditional random field parameterized by random forests, to estimate their local ancestry in the largest assembled admixed population to date, with 3,692 African Americans, 4,915 Asian Americans, and 3,777 Hispanics. The majority of MS-associated human leukocyte antigen (HLA) alleles, including the prominent HLA-DRB1*15:01 risk allele, exhibited cosmopolitan ancestry. Ancestry-specific MS-associated HLA alleles were also identified. Analysis of the HLA-DRB1*15:01 risk allele in African Americans revealed that alleles on the European haplotype conferred three times the disease risk compared to those on the African haplotype. Furthermore, we found evidence that the European and African HLA-DRB1*15:01 alleles exhibit single nucleotide polymorphism (SNP) differences in regions encoding the HLA-DRB1 antigen-binding heterodimer. Additional evidence for increased risk of MS conferred by the European haplotype were found for HLA-B*07:02 and HLA-A*03:01 in African Americans. Most of the 200 non-HLA MS SNPs previously established in European populations were not significantly associated with MS in admixed populations, nor were they ancestrally more European in cases compared to controls. Lastly, a genome-wide search of association between European ancestry and MS revealed a region of interest close to the ZNF596 gene on chromosome 8 in Hispanics; cases had a significantly higher proportion of European ancestry compared to controls. In conclusion, our study established that the genetic ancestry of MS-associated alleles is complex and implicated that difference in MS prevalence could be explained by the ancestry of MS-associated alleles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

