Epitranscriptomics: Correlation of N6-methyladenosine RNA methylation and pathway dysregulation in the hippocampus of HIV transgenic rats
- PMID: 30653517
- PMCID: PMC6336335
- DOI: 10.1371/journal.pone.0203566
Epitranscriptomics: Correlation of N6-methyladenosine RNA methylation and pathway dysregulation in the hippocampus of HIV transgenic rats
Abstract
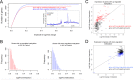
Internal RNA modifications have been known for decades, however their roles in mRNA regulation have only recently started to be elucidated. Here we investigated the most abundant mRNA modification, N6-methyladenosine (m6A) in transcripts from the hippocampus of HIV transgenic (Tg) rats. The distribution of m6A peaks within HIV transcripts in HIV Tg rats largely corresponded to the ones observed for HIV transcripts in cell lines and T cells. Host transcripts were found to be differentially m6A methylated in HIV Tg rats. The functional roles of the differentially m6A methylated pathways in HIV Tg rats is consistent with a key role of RNA methylation in the regulation of the brain transcriptome in chronic HIV disease. In particular, host transcripts show significant differential m6A methylation of genes involved in several pathways related to neural function, suggestive of synaptodendritic injury and neurodegeneration, inflammation and immune response, as well as RNA processing and metabolism, such as splicing. Changes in m6A methylation were usually positively correlated with differential expression, while differential m6A methylation of pathways involved in RNA processing were more likely to be negatively correlated with gene expression changes. Thus, sets of differentially m6A methylated, functionally-related transcripts appear to be involved in coordinated transcriptional responses in the context of chronic HIV. Altogether, our results support that m6A methylation represents an additional layer of regulation of HIV and host gene expression in vivo that contributes significantly to the transcriptional effects of chronic HIV.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

