Footprints of adaptive evolution revealed by whole Z chromosomes haplotypes in flycatchers
- PMID: 30653779
- PMCID: PMC6852393
- DOI: 10.1111/mec.15021
Footprints of adaptive evolution revealed by whole Z chromosomes haplotypes in flycatchers
Abstract
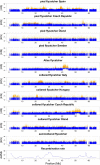
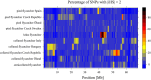
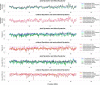
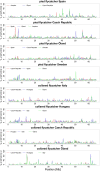
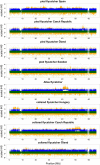
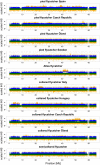
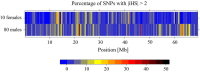
Detecting positive selection using genomic data is critical to understanding the role of adaptive evolution. Of particular interest in this context is sex chromosomes since they are thought to play a special role in local adaptation and speciation. We sought to circumvent the challenges associated with statistical phasing when using haplotype-based statistics in sweep scans by benefitting from that whole chromosome haplotypes of the sex chromosomes can be obtained by resequencing of individuals of the hemizygous sex. We analyzed whole Z chromosome haplotypes from 100 females from several populations of four black and white flycatcher species (in birds, females are ZW and males ZZ). Based on integrated haplotype score (iHS) and number of segregating sites by length (nSL) statistics, we found strong and frequent haplotype structure in several regions of the Z chromosome in each species. Most of these sweep signals were population-specific, with essentially no evidence for regions under selection shared among species. Some completed sweeps were revealed by the cross-population extended haplotype homozygosity (XP-EHH) statistic. Importantly, by using statistically phased Z chromosome data from resequencing of males, we failed to recover the signals of selection detected in analyses based on whole chromosome haplotypes from females; instead, what likely represent false signals of selection were frequently seen. This highlights the power issues in statistical phasing and cautions against conclusions from selection scans using such data. The detection of frequent selective sweeps on the avian Z chromosome supports a large role of sex chromosomes in adaptive evolution.
Keywords: Ficedula flycatchers; haplotype-based statistics; ongoing selection; sex chromosomes.
© 2019 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures
References
-
- Borge, T. , Lindroos, K. , Nádvorník, P. , Syvänen, A. , & Saetre, G. (2005). Amount of introgression in flycatcher hybrid zones reflects regional differences in pre and post‐zygotic barriers to gene exchange. Journal of Evolutionary Biology, 18, 1416–1424. 10.1111/j.1420-9101.2005.00964.x - DOI - PubMed
-
- Burri, R. , Nater, A. , Kawakami, T. , Mugal, C. F. , Olason, P. I. , Smeds, L. , … Ellegren, H. (2015). Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research, 25, 1656–1665. 10.1101/gr.196485.115 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

