A Rapid Method for Sequencing Double-Stranded RNAs Purified from Yeasts and the Identification of a Potent K1 Killer Toxin Isolated from Saccharomyces cerevisiae
- PMID: 30654470
- PMCID: PMC6356530
- DOI: 10.3390/v11010070
A Rapid Method for Sequencing Double-Stranded RNAs Purified from Yeasts and the Identification of a Potent K1 Killer Toxin Isolated from Saccharomyces cerevisiae
Abstract
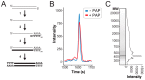
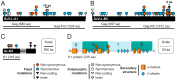
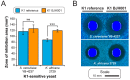
Mycoviruses infect a large number of diverse fungal species, but considering their prevalence, relatively few high-quality genome sequences have been determined. Many mycoviruses have linear double-stranded RNA genomes, which makes it technically challenging to ascertain their nucleotide sequence using conventional sequencing methods. Different specialist methodologies have been developed for the extraction of double-stranded RNAs from fungi and the subsequent synthesis of cDNAs for cloning and sequencing. However, these methods are often labor-intensive, time-consuming, and can require several days to produce cDNAs from double-stranded RNAs. Here, we describe a comprehensive method for the rapid extraction and sequencing of dsRNAs derived from yeasts, using short-read next generation sequencing. This method optimizes the extraction of high-quality double-stranded RNAs from yeasts and 3' polyadenylation for the initiation of cDNA synthesis for next-generation sequencing. We have used this method to determine the sequence of two mycoviruses and a double-stranded RNA satellite present within a single strain of the model yeast Saccharomyces cerevisiae. The quality and depth of coverage was sufficient to detect fixed and polymorphic mutations within viral populations extracted from a clonal yeast population. This method was also able to identify two fixed mutations within the alpha-domain of a variant K1 killer toxin encoded on a satellite double-stranded RNA. Relative to the canonical K1 toxin, these newly reported mutations increased the cytotoxicity of the K1 toxin against a specific species of yeast.
Keywords: dsRNA; killer toxin; mycovirus; sequencing; totivirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The prevalence of killer yeasts and double-stranded RNAs in the budding yeast Saccharomyces cerevisiae.FEMS Yeast Res. 2023 Jan 4;23:foad046. doi: 10.1093/femsyr/foad046. FEMS Yeast Res. 2023. PMID: 37935474 Free PMC article.
-
The Species-Specific Acquisition and Diversification of a K1-like Family of Killer Toxins in Budding Yeasts of the Saccharomycotina.PLoS Genet. 2021 Feb 4;17(2):e1009341. doi: 10.1371/journal.pgen.1009341. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33539346 Free PMC article.
-
Cloning and expression of a cDNA copy of the viral K28 killer toxin gene in yeast.Mol Gen Genet. 1995 Jan 20;246(2):236-46. doi: 10.1007/BF00294687. Mol Gen Genet. 1995. PMID: 7862095
-
Yeast dsRNA viruses: replication and killer phenotypes.Mol Microbiol. 1991 Oct;5(10):2331-8. doi: 10.1111/j.1365-2958.1991.tb02078.x. Mol Microbiol. 1991. PMID: 1665194 Review.
-
Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae.Annu Rev Microbiol. 1992;46:347-75. doi: 10.1146/annurev.mi.46.100192.002023. Annu Rev Microbiol. 1992. PMID: 1444259 Review.
Cited by
-
Mycoviruses: Past, Present, and Future.Viruses. 2019 Apr 19;11(4):361. doi: 10.3390/v11040361. Viruses. 2019. PMID: 31010228 Free PMC article.
-
A Simple Multiplex Reverse Transcription-PCR Method for the Diagnosis of L-A and M Totiviruses in Saccharomyces cerevisiae.Appl Environ Microbiol. 2022 Feb 22;88(4):e0221321. doi: 10.1128/AEM.02213-21. Epub 2021 Dec 15. Appl Environ Microbiol. 2022. PMID: 34910561 Free PMC article.
-
An optimized protocol for quality control of gene therapy vectors using nanopore direct RNA sequencing.Genome Res. 2024 Nov 20;34(11):1966-1975. doi: 10.1101/gr.279405.124. Genome Res. 2024. PMID: 39467647 Free PMC article.
-
The prevalence of killer yeasts and double-stranded RNAs in the budding yeast Saccharomyces cerevisiae.FEMS Yeast Res. 2023 Jan 4;23:foad046. doi: 10.1093/femsyr/foad046. FEMS Yeast Res. 2023. PMID: 37935474 Free PMC article.
-
Uncovering the Yeast Communities in Fungus-Growing Ant Colonies.Microb Ecol. 2023 Jul;86(1):624-635. doi: 10.1007/s00248-022-02099-1. Epub 2022 Aug 12. Microb Ecol. 2023. PMID: 35962280
References
-
- Milgroom M.G., Hillman B.I. In: Studies in Viral Ecology: Microbial and Botanical Host Systems. 1st ed. Hurst C.J., editor. Volume 1. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2011. pp. 217–253.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

