Phylodynamic Analysis of Ebola Virus Disease Transmission in Sierra Leone
- PMID: 30654482
- PMCID: PMC6356631
- DOI: 10.3390/v11010071
Phylodynamic Analysis of Ebola Virus Disease Transmission in Sierra Leone
Abstract
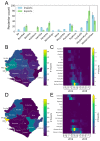
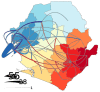
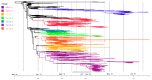
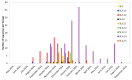
We generated genome sequences from 218 cases of Ebola virus disease (EVD) in Sierra Leone (SLE) during 2014⁻2015 to complement available datasets, particularly by including cases from a period of low sequence coverage during peak transmission of Ebola virus (EBOV) in the highly-affected Western Area division of SLE. The combined dataset was utilized to produce phylogenetic and phylodynamic inferences, to study sink⁻source dynamics and virus dispersal from highly-populated transmission hotspots. We identified four districts in SLE where EBOV was introduced and transmission occurred without onward exportation to other districts. We also identified six districts that substantially contributed to the dispersal of the virus and prolonged the EVD outbreak: five of these served as major hubs, with lots of movement in and out, and one acted primarily as a source, exporting the virus to other areas of the country. Positive correlations between case numbers, inter-district transition events, and district population sizes reaffirm that population size was a driver of EBOV transmission dynamics in SLE. The data presented here confirm the role of urban hubs in virus dispersal and of a delayed laboratory response in the expansion and perpetuation of the EVD outbreak in SLE.
Keywords: Ebola virus; Ebola virus disease; Filoviridae; Sierra Leone; West Africa; filovirus; phylodynamics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- World Health Organization Ebola Situation Report—30 March 2016. [(accessed on 3 May 2018)]; Available online: http://apps.who.int/ebola/current-situation/ebola-situation-report-30-ma....
-
- Park J.P., Dudas G., Wohl S., Goba A., Whitmer S.L., Andersen K.G., Sealfon R.S., Ladner J.T., Kugelman J.R., Matranga C.B., et al. Ebola virus epidemiology, transmission and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

