Comprehensive Analysis of Differentially Expressed Unigenes under NaCl Stress in Flax (Linum usitatissimum L.) Using RNA-Seq
- PMID: 30654562
- PMCID: PMC6359340
- DOI: 10.3390/ijms20020369
Comprehensive Analysis of Differentially Expressed Unigenes under NaCl Stress in Flax (Linum usitatissimum L.) Using RNA-Seq
Abstract
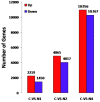
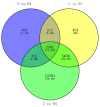
Flax (Linum usitatissimum L.) is an important industrial crop that is often cultivated on marginal lands, where salt stress negatively affects yield and quality. High-throughput RNA sequencing (RNA-seq) using the powerful Illumina platform was employed for transcript analysis and gene discovery to reveal flax response mechanisms to salt stress. After cDNA libraries were constructed from flax exposed to water (negative control) or salt (100 mM NaCl) for 12 h, 24 h or 48 h, transcription expression profiles and cDNA sequences representing expressed mRNA were obtained. A total of 431,808,502 clean reads were assembled to form 75,961 unigenes. After ruling out short-length and low-quality sequences, 33,774 differentially expressed unigenes (DEUs) were identified between salt-stressed and unstressed control (C) flax. Of these DEUs, 3669, 8882 and 21,223 unigenes were obtained from flax exposed to salt for 12 h (N1), 24 h (N2) and 48 h (N4), respectively. Gene function classification and pathway assignments of 2842 DEUs were obtained by comparing unigene sequences to information within public data repositories. qRT-PCR of selected DEUs was used to validate flax cDNA libraries generated for various durations of salt exposure. Based on transcriptome sequences, 1777 EST-SSRs were identified of which trinucleotide and dinucleotide repeat microsatellite motifs were most abundant. The flax DEUs and EST-SSRs identified here will serve as a powerful resource to better understand flax response mechanisms to salt exposure for development of more salt-tolerant varieties of flax.
Keywords: DEUs; EST-SSR; NaCl stress; RNA-seq; flax.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
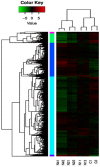
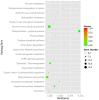

References
-
- Yu Y., Huang W.G., Chen H.Y., Wu G.W., Yuan H.M., Song X.X., Kang Q.H., Zhao D.S., Jiang W.D., Liu Y., et al. Identification of differentially expressed genes in flax (Linum usitatissimum L.) under saline-alkaline stress by digital gene expression. Gene. 2014;549:113–122. doi: 10.1016/j.gene.2014.07.053. - DOI - PubMed
-
- El-Hariri D.M., Al-Kordy M.A., Hassanein M.S., Ahmed M.A. Partition of photosynthates and energy production in different flax cultivars. J. Nat. Fibers. 2005 doi: 10.1300/J395v01n04_01. - DOI
-
- Pecenka R., Fürll C., Ola D.C., Budde J., Gusovius H.J. Efficient use of agricultural land in production of energy: Natural insulation versus bio-energy; Proceedings of the International Conference of Agricultural Engineering CIGR-AgEng, Agriculture and Engineering for a Healthier Life; Valencia, Spain. 8–12 July 2012.
-
- Marshall G. Flax: Breeding and utilisation. Plant Growth Regul. 1991;10:171–172. doi: 10.1007/BF00024965. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

