Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication
- PMID: 30654597
- PMCID: PMC6357182
- DOI: 10.3390/v11010073
Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication
Abstract
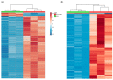
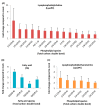
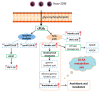
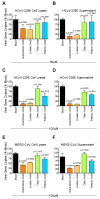
Lipids play numerous indispensable cellular functions and are involved in multiple steps in the replication cycle of viruses. Infections by human-pathogenic coronaviruses result in diverse clinical outcomes, ranging from self-limiting flu-like symptoms to severe pneumonia with extrapulmonary manifestations. Understanding how cellular lipids may modulate the pathogenicity of human-pathogenic coronaviruses remains poor. To this end, we utilized the human coronavirus 229E (HCoV-229E) as a model coronavirus to comprehensively characterize the host cell lipid response upon coronavirus infection with an ultra-high performance liquid chromatography-mass spectrometry (UPLC⁻MS)-based lipidomics approach. Our results revealed that glycerophospholipids and fatty acids (FAs) were significantly elevated in the HCoV-229E-infected cells and the linoleic acid (LA) to arachidonic acid (AA) metabolism axis was markedly perturbed upon HCoV-229E infection. Interestingly, exogenous supplement of LA or AA in HCoV-229E-infected cells significantly suppressed HCoV-229E virus replication. Importantly, the inhibitory effect of LA and AA on virus replication was also conserved for the highly pathogenic Middle East respiratory syndrome coronavirus (MERS-CoV). Taken together, our study demonstrated that host lipid metabolic remodeling was significantly associated with human-pathogenic coronavirus propagation. Our data further suggested that lipid metabolism regulation would be a common and druggable target for coronavirus infections.
Keywords: HCoV-229E; MERS-CoV; UHPLC–MS; lipidomics.
Conflict of interest statement
J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited, and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. The other authors declared no conflict of interest. The funding sources had no role in study design, data collection, analysis or interpretation or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

