UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond
- PMID: 30655309
- PMCID: PMC6426291
- DOI: 10.1261/rna.070136.118
UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond
Abstract
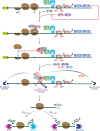
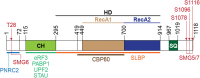
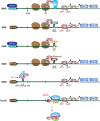
Nonsense-mediated mRNA decay (NMD), which is arguably the best-characterized translation-dependent regulatory pathway in mammals, selectively degrades mRNAs as a means of post-transcriptional gene control. Control can be for the purpose of ensuring the quality of gene expression. Alternatively, control can facilitate the adaptation of cells to changes in their environment. The key to NMD, no matter what its purpose, is the ATP-dependent RNA helicase upstream frameshift 1 (UPF1), without which NMD fails to occur. However, UPF1 does much more than regulate NMD. As examples, UPF1 is engaged in functionally diverse mRNA decay pathways mediated by a variety of RNA-binding proteins that include staufen, stem-loop-binding protein, glucocorticoid receptor, and regnase 1. Moreover, UPF1 promotes tudor-staphylococcal/micrococcal-like nuclease-mediated microRNA decay. In this review, we first focus on how the NMD machinery recognizes an NMD target and triggers mRNA degradation. Next, we compare and contrast the mechanisms by which UPF1 functions in the decay of other mRNAs and also in microRNA decay. UPF1, as a protein polymath, engenders cells with the ability to shape their transcriptome in response to diverse biological and physiological needs.
Keywords: Staufen-mediated mRNA decay; UPF1; nonsense-mediated mRNA decay.
© 2019 Kim and Maquat; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
