Virulence and genomic features of a bla CTX-M-3 and bla CTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65
- PMID: 30655681
- PMCID: PMC6322562
- DOI: 10.2147/IDR.S187289
Virulence and genomic features of a bla CTX-M-3 and bla CTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65
Abstract
Background: Capsular serotype K2 Klebsiella pneumoniae of sequence type (ST) 65 has been recognized as a hypervirulent clone. Simultaneous presence of different bla CTX-M genes has never been reported in this clone. In the present study, the genetic characteristics and virulence phenotype of a CTX-M-3 and CTX-M-14 coproducing ST65 K. pneumoniae human isolate, KP_06, that caused an intracranial infection, are evaluated.
Methods: The potential virulence of KP_06 was assayed by in vitro and in vivo methods. The molecular biology and whole-genome sequencing technology were used to analyze the genomic features associated with the virulence of this strain.
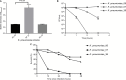
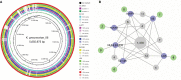
Results: The KP_06 exhibited typical features of hypervirulent K. pneumoniae (hvKP), showing hypermucoviscosity phenotype and belonging to K2 and ST65. Apart from virulence genes linked to hvKP, including rmpA, rmpA2, and clb cluster and genes encoding siderophores, it was found to harbor a ~170 kb pLVPK-like virulence plasmid. In contrast to most hvKP, KP_06 was resistant to cephalosporins and the coexistence of bla CTX-M-3 and bla CTX-M-14 was detected. Further experiments demonstrated that this strain was classified as a nonbiofilm producer and serum sensitivity (grade 1) and killed only 30% of Galleria mellonella inoculated with 1×106 colony-forming unit of the specimen within 48 hours, suggesting relatively low virulence. Comparative genomic analysis of KP_06 with five K2 hypermucoviscous K. pneumoniae (HMKP) revealed seven unique orthologies with varied function in this strain. Intriguingly, the virulence genes identified in KP-06 were unexpectedly more diverse than those observed in five other K2 HMKP strains.
Conclusion: Our data support the notion that neither virulence-associated genes (clusters) nor the pLVPK-like virulence plasmid is sufficient for the hypervirulence of K. pneumoniae. Future studies aiming to explore the virulence of K. pneumoniae should take genome-based profile together with experimental work. The detailed mechanism involving in the impaired virulence of KP_06 remains to be further explored.
Keywords: Klebsiella pneumoniae; ST65; blaCTX-M; comparative genome; serotype K2; virulence factor.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures
References
-
- Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. - PubMed
LinkOut - more resources
Full Text Sources

