Identification of hub genes and pathways in glioblastoma by bioinformatics analysis
- PMID: 30655863
- PMCID: PMC6312941
- DOI: 10.3892/ol.2018.9644
Identification of hub genes and pathways in glioblastoma by bioinformatics analysis
Abstract
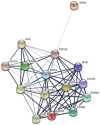
Glioblastoma (GBM) is the most common type of malignant brain tumor, and is associated with poor patient prognosis. A comprehensive understanding of the molecular mechanism underlying GBM may help to guide the identification of novel diagnoses and treatment targets. The gene expression profile of the GSE4290 GBM dataset was analyzed in order to identify differentially expressed genes (DEGs). Enriched pathways were identified through Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes analyses. A protein-protein interaction network was constructed in order to identify hub genes and for module analysis. Expression and survival analyses were conducted in order to screen and validate critical genes. A total of 1,801 DEGs were recorded, including 620 upregulated and 1,181 downregulated genes. Upregulated DEGs were enriched in the terms 'mitotic cell cycle process', 'mitotic cell cycle' and 'cell cycle process'. Downregulated genes were enriched in 'transsynaptic signaling', 'anterograde transsynaptic signaling' and 'synaptic signaling'. A total of 15 hub genes, which displayed a high degree of connectivity, were selected. These genes included vascular endothelial growth factor A, cyclin-dependent kinase 1 (CDK1), cell-division cycle protein 20 (CDC20), aurora kinase A (AURKA), and budding uninhibited by benzimidazoles 1 (BUB1). The identified DEGs and hub genes may help guide investigations on the mechanisms underlying the development and progression of GBM. CDK1, CDC20, AURKA and BUB1, which are involved in cell cycle pathways, may be potential targets in the diagnosis and therapy of GBM.
Keywords: bioinformatics analysis; glioblastoma; module analysis; pathway analysis; protein-protein interaction.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
