Lugdunomycin, an Angucycline-Derived Molecule with Unprecedented Chemical Architecture
- PMID: 30656821
- PMCID: PMC6519343
- DOI: 10.1002/anie.201814581
Lugdunomycin, an Angucycline-Derived Molecule with Unprecedented Chemical Architecture
Abstract
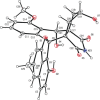
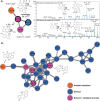
The angucyclines form the largest family of polycyclic aromatic polyketides, and have been studied extensively. Herein, we report the discovery of lugdunomycin, an angucycline-derived polyketide, produced by Streptomyces species QL37. Lugdunomycin has unique structural characteristics, including a heptacyclic ring system, a spiroatom, two all-carbon stereocenters, and a benzaza-[4,3,3]propellane motif. Considering the structural novelty, we propose that lugdunomycin represents a novel subclass of aromatic polyketides. Metabolomics, combined with MS-based molecular networking analysis of Streptomyces sp. QL37, elucidated 24 other rearranged and non-rearranged angucyclines, 11 of which were previously undescribed. A biosynthetic route for the lugdunomycin and limamycins is also proposed. This work demonstrates that revisiting well-known compound families and their producer strains still is a promising approach for drug discovery.
Keywords: Baeyer-Villiger oxidation; angucycline; molecular networking; natural product; polyketide.
© 2019 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Antibacterial Pentacyclic Polyketides from a Soil-Derived Streptomyces.J Nat Prod. 2020 Jun 26;83(6):1919-1924. doi: 10.1021/acs.jnatprod.0c00161. Epub 2020 Jun 10. J Nat Prod. 2020. PMID: 32519857
-
Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.ACS Chem Biol. 2020 Sep 18;15(9):2529-2538. doi: 10.1021/acschembio.0c00564. Epub 2020 Sep 8. ACS Chem Biol. 2020. PMID: 32840360 Free PMC article.
-
Molecular basis of dimer formation during the biosynthesis of benzofluorene-containing atypical angucyclines.Nat Commun. 2018 May 25;9(1):2088. doi: 10.1038/s41467-018-04487-z. Nat Commun. 2018. PMID: 29802272 Free PMC article.
-
The Chemistry of Angucyclines.Chempluschem. 2024 Oct;89(10):e202400307. doi: 10.1002/cplu.202400307. Epub 2024 Aug 21. Chempluschem. 2024. PMID: 38958029 Review.
-
[Novel angucycline/angucyclinone family of natural products discovered between 2010 and 2020].Sheng Wu Gong Cheng Xue Bao. 2021 Jun 25;37(6):2147-2165. doi: 10.13345/j.cjb.210033. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 34227300 Review. Chinese.
Cited by
-
Ring D-Modified and Highly Reduced Angucyclinones From Marine Sediment-Derived Streptomyces sp.Front Chem. 2021 Oct 12;9:756962. doi: 10.3389/fchem.2021.756962. eCollection 2021. Front Chem. 2021. PMID: 34712650 Free PMC article.
-
Donghaecyclinones A-C: New Cytotoxic Rearranged Angucyclinones from a Volcanic Island-Derived Marine Streptomyces sp.Mar Drugs. 2020 Feb 18;18(2):121. doi: 10.3390/md18020121. Mar Drugs. 2020. PMID: 32085561 Free PMC article.
-
Unravelling key enzymatic steps in C-ring cleavage during angucycline biosynthesis.Commun Chem. 2023 Dec 18;6(1):281. doi: 10.1038/s42004-023-01059-1. Commun Chem. 2023. PMID: 38110491 Free PMC article.
-
Genome-based classification of Streptomyces pinistramenti sp. nov., a novel actinomycete isolated from a pine forest soil in Poland with a focus on its biotechnological and ecological properties.Antonie Van Leeuwenhoek. 2022 Jun;115(6):783-800. doi: 10.1007/s10482-022-01734-8. Epub 2022 Apr 11. Antonie Van Leeuwenhoek. 2022. PMID: 35404014
-
The ubiquitous catechol moiety elicits siderophore and angucycline production in Streptomyces.Commun Chem. 2022 Feb 3;5(1):14. doi: 10.1038/s42004-022-00632-4. Commun Chem. 2022. PMID: 36697563 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

