Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus
- PMID: 30656859
- PMCID: PMC6854852
- DOI: 10.1002/mbo3.791
Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus
Abstract
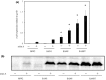
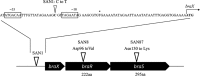
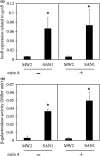
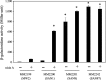
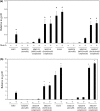
Nisin A is a lantibiotic produced by Lactococcus lactis that is widely used as a food preservative. In Staphylococcus aureus, the BraRS two-component system (TCS) senses nisin A and regulates the expression of the ABC transporter VraDE, which is responsible for nisin A resistance. In this study, we exposed S. aureus to a sub-minimum inhibition concentration of nisin A and obtained three spontaneous mutants that were highly resistant to this lantibiotic, designated as SAN (S. aureus nisin resistant) 1, SAN8, and SAN87. In the wild-type S. aureus strain, VraDE expression was induced by nisin A. In contrast, SAN8 and SAN87 showed constitutively high VraDE expression, even in the absence of nisin A, while SAN1 showed higher BraRS expression, which resulted in high VraDE expression in the presence of nisin A. We identified a single mutation in the promoter region of braXRS in SAN1, whereas SAN8 and SAN87 had single mutations in braR and braS, respectively. Interestingly, even the unphosphorylated form of the mutant BraR protein induced VraDE expression. These results indicate that conformational changes in BraS or BraR resulting from the point mutations may result in the constitutive expression of VraDE, allowing S. aureus to adapt to high concentrations of nisin A.
Keywords: ABC transporter; mutation; nisin A; resistance; two-component system.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Bal, A. M. , Coombs, G. W. , Holden, M. T. G. , Lindsay, J. A. , Nimmo, G. R. , Tattevin, P. , & Skov, R. L. (2016). Genomic insights into the emergence and spread of international clones of healthcare‐, community‐ and livestock‐associated meticillin‐resistant Staphylococcus aureus: Blurring of the traditional definitions. Journal of Global Antimicrobial Resistance, 6, 95–101. 10.1016/j.jgar.2016.04.004 - DOI - PubMed
-
- Bierbaum, G. , & Sahl, H. G. (2009). Lantibiotics: Mode of action, biosynthesis and bioengineering. Current Pharmaceutical Biotechnology, 10, 2–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

