Analyses of four new Caulobacter Phicbkviruses indicate independent lineages
- PMID: 30657445
- PMCID: PMC7011694
- DOI: 10.1099/jgv.0.001218
Analyses of four new Caulobacter Phicbkviruses indicate independent lineages
Abstract
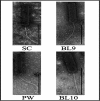
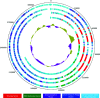
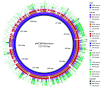
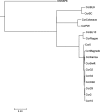
Bacteriophages with genomes larger than 200 kbp are considered giant phages, and the giant Phicbkviruses are the most frequently isolated Caulobacter crescentus phages. In this study, we compare six bacteriophage genomes that differ from the genomes of the majority of Phicbkviruses. Four of these genomes are much larger than those of the rest of the Phicbkviruses, with genome sizes that are more than 250 kbp. A comparison of 16 Phicbkvirus genomes identified a 'core genome' of 69 genes that is present in all of these Phicbkvirus genomes, as well as shared accessory genes and genes that are unique for each phage. Most of the core genes are clustered into the regions coding for structural proteins or those involved in DNA replication. A phylogenetic analysis indicated that these 16 CaulobacterPhicbkvirus genomes are related, but they represent four distinct branches of the Phicbkvirus genomic tree with distantly related branches sharing little nucleotide homology. In contrast, pairwise comparisons within each branch of the phylogenetic tree showed that more than 80 % of the entire genome is shared among phages within a group. This conservation of the genomes within each branch indicates that horizontal gene transfer events between the groups are rare. Therefore, the Phicbkvirus genus consists of at least four different phylogenetic branches that are evolving independently from one another. One of these branches contains a 27-gene inversion relative to the other three branches. Also, an analysis of the tRNA genes showed that they are relatively mobile within the Phicbkvirus genus.
Keywords: Bacteriophage; Caulobacter; DNA virus; genomics; pan-genome.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

