Expression Changes of Structural Protein Genes May Be Related to Adaptive Skin Characteristics Specific to Humans
- PMID: 30657921
- PMCID: PMC6402313
- DOI: 10.1093/gbe/evz007
Expression Changes of Structural Protein Genes May Be Related to Adaptive Skin Characteristics Specific to Humans
Abstract
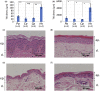
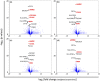
Human skin is morphologically and physiologically different from the skin of other primates. However, the genetic causes underlying human-specific skin characteristics remain unclear. Here, we quantitatively demonstrate that the epidermis and dermis of human skin are significantly thicker than those of three Old World monkey species. In addition, we indicate that the topography of the epidermal basement membrane zone shows a rete ridge in humans but is flat in the Old World monkey species examined. Subsequently, we comprehensively compared gene expression levels between human and nonhuman great ape skin using next-generation cDNA sequencing (RNA-Seq). We identified four structural protein genes associated with the epidermal basement membrane zone or elastic fibers in the dermis (COL18A1, LAMB2, CD151, and BGN) that were expressed significantly greater in humans than in nonhuman great apes, suggesting that these differences may be related to the rete ridge and rich elastic fibers present in human skin. The rete ridge may enhance the strength of adhesion between the epidermis and dermis in skin. This ridge, along with a thick epidermis and rich elastic fibers might contribute to the physical strength of human skin with a low amount of hair. To estimate transcriptional regulatory regions for COL18A1, LAMB2, CD151, and BGN, we examined conserved noncoding regions with histone modifications that can activate transcription in skin cells. Human-specific substitutions in these regions, especially those located in binding sites of transcription factors which function in skin, may alter the gene expression patterns and give rise to the human-specific adaptive skin characteristics.
Keywords: elastic fibers; epidermal basement membrane zone; primates; rete ridge; transcriptional regulatory region.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Baggerly KA, Deng L, Morris JS, Aldaz CM.. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 1912:1477–1483. - PubMed
-
- Barski A, et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 1294:823–837. - PubMed
-
- Bianco P, Fisher LW, Young MF, Termine JD, Robey PG.. 1990. Expression and localization of the two small proteoglycans biglycan and decorin in developing human skeletal and non-skeletal tissues. J Histochem Cytochem. 3811:1549–1563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

