Targeted Genomic Profiling of Acral Melanoma
- PMID: 30657954
- PMCID: PMC6792090
- DOI: 10.1093/jnci/djz005
Targeted Genomic Profiling of Acral Melanoma
Abstract
Background: Acral melanoma is a rare type of melanoma that affects world populations irrespective of skin color and has worse survival than other cutaneous melanomas. It has relatively few single nucleotide mutations without the UV signature of cutaneous melanomas, but instead has a genetic landscape characterized by structural rearrangements and amplifications. BRAF mutations are less common than in other cutaneous melanomas, and knowledge about alternative therapeutic targets is incomplete.
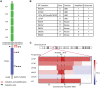
Methods: To identify alternative therapeutic targets, we performed targeted deep-sequencing on 122 acral melanomas. We confirmed the loss of the tumor suppressors p16 and NF1 by immunohistochemistry in select cases.
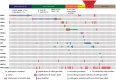
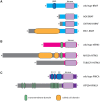
Results: In addition to BRAF (21.3%), NRAS (27.9%), and KIT (11.5%) mutations, we identified a broad array of MAPK pathway activating alterations, including fusions of BRAF (2.5%), NTRK3 (2.5%), ALK (0.8%), and PRKCA (0.8%), which can be targeted by available inhibitors. Inactivation of NF1 occurred in 18 cases (14.8%). Inactivation of the NF1 cooperating factor SPRED1 occurred in eight cases (6.6%) as an alternative mechanism of disrupting the negative regulation of RAS. Amplifications recurrently affected narrow loci containing PAK1 and GAB2 (n = 27, 22.1%), CDK4 (n = 27, 22.1%), CCND1 (n = 24, 19.7%), EP300 (n = 20, 16.4%), YAP1 (n = 15, 12.3%), MDM2 (n = 13, 10.7%), and TERT (n = 13, 10.7%) providing additional and possibly complementary therapeutic targets. Acral melanomas with BRAFV600E mutations harbored fewer genomic amplifications and were more common in patients with European ancestry.
Conclusion: Our findings support a new, molecularly based subclassification of acral melanoma with potential therapeutic implications: BRAFV600E mutant acral melanomas with characteristics similar to nonacral melanomas that could benefit from BRAF inhibitor therapy, and non-BRAFV600E mutant acral melanomas. Acral melanomas without BRAFV600E mutations harbor a broad array of therapeutically relevant alterations. Expanded molecular profiling would increase the detection of potentially targetable alterations for this subtype of acral melanoma.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Desai A, Ugorji R, Khachemoune A.. Acral melanoma foot lesions. Part 1: epidemiology, aetiology, and molecular pathology. Clin Exp Dermatol. 2017;428:845–848. - PubMed
-
- Durbec F, Martin L, Derancourt C, Grange F.. Melanoma of the hand and foot: epidemiological, prognostic and genetic features. A systematic review. Br J Dermatol. 2012;1664:727–739. - PubMed
-
- Bello DM, Chou JF, Panageas KS.. Prognosis of acral melanoma: a series of 281 patients. Ann Surg Oncol. 2013;2011:3618–3625. - PubMed
-
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;35320:2135–2147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

