In-Cell NMR: Analysis of Protein-Small Molecule Interactions, Metabolic Processes, and Protein Phosphorylation
- PMID: 30658393
- PMCID: PMC6359726
- DOI: 10.3390/ijms20020378
In-Cell NMR: Analysis of Protein-Small Molecule Interactions, Metabolic Processes, and Protein Phosphorylation
Abstract
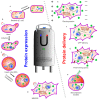
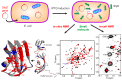
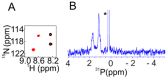
Nuclear magnetic resonance (NMR) spectroscopy enables the non-invasive observation of biochemical processes, in living cells, at comparably high spectral and temporal resolution. Preferably, means of increasing the detection limit of this powerful analytical method need to be applied when observing cellular processes under physiological conditions, due to the low sensitivity inherent to the technique. In this review, a brief introduction to in-cell NMR, protein-small molecule interactions, posttranslational phosphorylation, and hyperpolarization NMR methods, used for the study of metabolites in cellulo, are presented. Recent examples of method development in all three fields are conceptually highlighted, and an outlook into future perspectives of this emerging area of NMR research is given.
Keywords: DNP; in-cell NMR; in-situ NMR; protein NMR; review.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
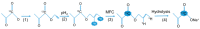

Similar articles
-
NMR spectroscopy for metabolomics in the living system: recent progress and future challenges.Anal Bioanal Chem. 2024 Apr;416(9):2319-2334. doi: 10.1007/s00216-024-05137-8. Epub 2024 Jan 19. Anal Bioanal Chem. 2024. PMID: 38240793 Free PMC article. Review.
-
NMR-based Stable Isotope Resolved Metabolomics in systems biochemistry.Arch Biochem Biophys. 2017 Aug 15;628:123-131. doi: 10.1016/j.abb.2017.02.009. Epub 2017 Mar 2. Arch Biochem Biophys. 2017. PMID: 28263717 Free PMC article. Review.
-
Polarization Transfer from Ligands Hyperpolarized by Dissolution Dynamic Nuclear Polarization for Screening in Drug Discovery.ChemMedChem. 2015 Sep;10(9):1559-63. doi: 10.1002/cmdc.201500241. Epub 2015 Aug 4. ChemMedChem. 2015. PMID: 26315550
-
NMR in structure-based drug design.Essays Biochem. 2017 Nov 8;61(5):485-493. doi: 10.1042/EBC20170037. Print 2017 Nov 8. Essays Biochem. 2017. PMID: 29118095 Review.
-
Applications of NMR spectroscopy to systems biochemistry.Prog Nucl Magn Reson Spectrosc. 2016 Feb;92-93:18-53. doi: 10.1016/j.pnmrs.2016.01.005. Epub 2016 Feb 6. Prog Nucl Magn Reson Spectrosc. 2016. PMID: 26952191 Free PMC article. Review.
Cited by
-
High-Resolution Magic Angle Spinning (HR-MAS) NMR-Based Fingerprints Determination in the Medicinal Plant Berberis laurina.Molecules. 2020 Aug 11;25(16):3647. doi: 10.3390/molecules25163647. Molecules. 2020. PMID: 32796509 Free PMC article.
-
Molecular interactions between a diphenyl scaffold and PED/PEA15: Implications for type II diabetes therapeutics targeting PED/PEA15 - Phospholipase D1 interaction.Comput Struct Biotechnol J. 2024 May 4;23:2001-2010. doi: 10.1016/j.csbj.2024.04.063. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38770160 Free PMC article.
-
Label-free protein-structure-sensitive live-cell microscopy for patient-specific assessment of myeloma therapy.Nat Biomed Eng. 2025 Jul 14. doi: 10.1038/s41551-025-01443-3. Online ahead of print. Nat Biomed Eng. 2025. PMID: 40659832
-
Folding and Stability of Ankyrin Repeats Control Biological Protein Function.Biomolecules. 2021 Jun 5;11(6):840. doi: 10.3390/biom11060840. Biomolecules. 2021. PMID: 34198779 Free PMC article. Review.
-
Challenges and Advances in the Application of Dynamic Nuclear Polarization to Liquid-State NMR Spectroscopy.J Phys Chem B. 2021 May 27;125(20):5171-5190. doi: 10.1021/acs.jpcb.0c10937. Epub 2021 May 7. J Phys Chem B. 2021. PMID: 33960784 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

