Colorectal Cancer Prognosis is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study
- PMID: 30658510
- PMCID: PMC6351956
- DOI: 10.3390/jcm8010111
Colorectal Cancer Prognosis is Not Associated with BRAF and KRAS Mutations-A STROBE Compliant Study
Abstract
Background: We investigated the associations between v-Raf murine sarcoma viral oncogene homolog B1 (BRAFV600E, henceforth BRAF) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations and colorectal cancer (CRC) prognosis, using The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GSE39582) datasets.
Materials and methods: The effects of BRAF and KRAS mutations on overall survival (OS) and disease-free survival (DFS) of CRC were evaluated.
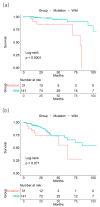
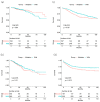
Results: The mutational status of BRAF and KRAS genes was not associated with overall survival (OS) or DFS of the CRC patients drawn from the TCGA database. The 3-year OS and DFS rates of the BRAF mutation (+) vs. mutation (-) groups were 92.6% vs. 90.4% and 79.7% vs. 68.4%, respectively. The 3-year OS and DFS rates of the KRAS mutation (+) vs. mutation (-) groups were 90.4% vs. 90.5% and 65.3% vs. 73.5%, respectively. In stage II patients, however, the 3-year OS rate was lower in the BRAF mutation (+) group than in the mutation (-) group (85.5% vs. 97.7%, p <0.001). The mutational status of BRAF genes of 497 CRC patients drawn from the GSE39582 database was not associated with OS or DFS. The 3-year OS and DFS rates of BRAF mutation (+) vs. mutation (-) groups were 75.7% vs. 78.9% and 73.6% vs. 71.1%, respectively. However, KRAS mutational status had an effect on 3-year OS rate (71.9% mutation (+) vs. 83% mutation (-), p = 0.05) and DFS rate (66.3% mutation (+) vs. 74.6% mutation (-), p = 0.013).
Conclusions: We found no consistent association between the mutational status of BRAF nor KRAS and the OS and DFS of CRC patients from the TCGA and GSE39582 databases. Studies with longer-term records and larger patient numbers may be necessary to expound the influence of BRAF and KRAS mutations on the outcomes of CRC.
Keywords: BRAF; KRAS; colorectal cancer; disease-free survival; overall survival.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer.JAMA Surg. 2018 Jul 18;153(7):e180996. doi: 10.1001/jamasurg.2018.0996. Epub 2018 Jul 18. JAMA Surg. 2018. PMID: 29799910 Free PMC article.
-
Prognostic Effect of BRAF and KRAS Mutations in Patients With Stage III Colon Cancer Treated With Leucovorin, Fluorouracil, and Oxaliplatin With or Without Cetuximab: A Post Hoc Analysis of the PETACC-8 Trial.JAMA Oncol. 2016 May 1;2(5):643-653. doi: 10.1001/jamaoncol.2015.5225. JAMA Oncol. 2016. PMID: 26768652
-
The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients.BMC Cancer. 2017 Jun 5;17(1):403. doi: 10.1186/s12885-017-3381-7. BMC Cancer. 2017. PMID: 28583095 Free PMC article.
-
BRAF and KRAS mutations in metastatic colorectal cancer: future perspectives for personalized therapy.Gastroenterol Rep (Oxf). 2020 Jun 15;8(3):192-205. doi: 10.1093/gastro/goaa022. eCollection 2020 Jun. Gastroenterol Rep (Oxf). 2020. PMID: 32665851 Free PMC article. Review.
-
Clinical implications of BRAF mutation test in colorectal cancer.Gastroenterol Hepatol Bed Bench. 2013 Winter;6(1):6-13. Gastroenterol Hepatol Bed Bench. 2013. PMID: 24834238 Free PMC article. Review.
Cited by
-
Histology and its prognostic effect on KRAS-mutated colorectal carcinomas in Korea.Oncol Lett. 2020 Jul;20(1):655-666. doi: 10.3892/ol.2020.11606. Epub 2020 May 13. Oncol Lett. 2020. PMID: 32565990 Free PMC article.
-
Upregulation of SLC2A3 gene and prognosis in colorectal carcinoma: analysis of TCGA data.BMC Cancer. 2019 Apr 3;19(1):302. doi: 10.1186/s12885-019-5475-x. BMC Cancer. 2019. PMID: 30943948 Free PMC article.
-
Identification of Hub Genes Related to Carcinogenesis and Prognosis in Colorectal Cancer Based on Integrated Bioinformatics.Mediators Inflamm. 2020 Apr 9;2020:5934821. doi: 10.1155/2020/5934821. eCollection 2020. Mediators Inflamm. 2020. PMID: 32351322 Free PMC article.
-
Identification of ATP8B1 as a Tumor Suppressor Gene for Colorectal Cancer and Its Involvement in Phospholipid Homeostasis.Biomed Res Int. 2020 Sep 29;2020:2015648. doi: 10.1155/2020/2015648. eCollection 2020. Biomed Res Int. 2020. PMID: 33062669 Free PMC article.
-
KRAS and BRAF Mutations as Prognostic and Predictive Biomarkers for Standard Chemotherapy Response in Metastatic Colorectal Cancer: A Single Institutional Study.Cells. 2020 Jan 15;9(1):219. doi: 10.3390/cells9010219. Cells. 2020. PMID: 31952366 Free PMC article.
References
-
- Amin M.B., Edge S., Greene F., Byrd D.R., Brookland R.K., Washington M.K., Gershenwald J.E., Compton C.C., Hess K.R., Sullivan D.C., et al. AJCC Cancer Staging Manual. 8th ed. Springer; New York, NY, USA: 2017. pp. XVII, 1032.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

