Proteomic analysis reveals response of differential wheat (Triticum aestivum L.) genotypes to oxygen deficiency stress
- PMID: 30658567
- PMCID: PMC6339445
- DOI: 10.1186/s12864-018-5405-3
Proteomic analysis reveals response of differential wheat (Triticum aestivum L.) genotypes to oxygen deficiency stress
Abstract
Background: Waterlogging is one of the main abiotic stresses that limit wheat production. Quantitative proteomics analysis has been applied in the study of crop abiotic stress as an effective way in recent years (e.g. salt stress, drought stress, heat stress and waterlogging stress). However, only a few proteins related to primary metabolism and signal transduction, such as UDP - glucose dehydrogenase, UGP, beta glucosidases, were reported to response to waterlogging stress in wheat. The differentially expressed proteins between genotypes of wheat in response to waterlogging are less-defined. In this study, two wheat genotypes, one is sensitive to waterlogging stress (Seri M82, named as S) and the other is tolerant to waterlogging (CIGM90.863, named as T), were compared in seedling roots under hypoxia conditions to evaluate the different responses at proteomic level.
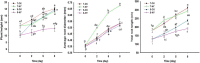
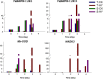
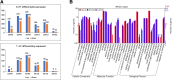
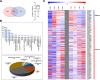
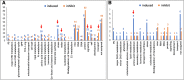
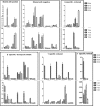
Results: A total of 4560 proteins were identified and the number of differentially expressed proteins (DEPs) were 361, 640, 788 in S and 33, 207, 279 in T in 1, 2, 3 days, respectively. These DEPs included 270 common proteins, 681 S-specific and 50 T-specific proteins, most of which were misc., protein processing, DNA and RNA processing, amino acid metabolism and stress related proteins induced by hypoxia. Some specific proteins related to waterlogging stress, including acid phosphatase, oxidant protective enzyme, S-adenosylmethionine synthetase 1, were significantly different between S and T. A total of 20 representative genes encoding DEPs, including 7 shared DEPs and 13 cultivar-specific DEPs, were selected for further RT-qPCR analysis. Fourteen genes showed consistent dynamic expression patterns at mRNA and protein levels.
Conclusions: Proteins involved in primary metabolisms and protein processing were inclined to be affected under hypoxia stress. The negative effects were more severe in the sensitive genotype. The expression patterns of some specific proteins, such as alcohol dehydrogenases and S-adenosylmethionine synthetase 1, could be applied as indexes for improving the waterlogging tolerance in wheat. Some specific proteins identified in this study will facilitate the subsequent protein function validation and biomarker development.
Keywords: Hypoxic stress; Proteomics; Triticum aestivum L.; Waterlogging tolerance.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests” in this section.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat.Mol Cell Proteomics. 2009 Dec;8(12):2676-86. doi: 10.1074/mcp.M900052-MCP200. Epub 2009 Sep 3. Mol Cell Proteomics. 2009. PMID: 19734139 Free PMC article.
-
Label-free quantitative proteomic analysis of drought stress-responsive late embryogenesis abundant proteins in the seedling leaves of two wheat (Triticum aestivum L.) genotypes.J Proteomics. 2018 Feb 10;172:122-142. doi: 10.1016/j.jprot.2017.09.016. Epub 2017 Oct 2. J Proteomics. 2018. PMID: 28982538
-
Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration.BMC Plant Biol. 2016 Aug 31;16(1):188. doi: 10.1186/s12870-016-0871-8. BMC Plant Biol. 2016. PMID: 27576435 Free PMC article.
-
The wheat chloroplastic proteome.J Proteomics. 2013 Nov 20;93:326-42. doi: 10.1016/j.jprot.2013.03.009. Epub 2013 Apr 3. J Proteomics. 2013. PMID: 23563086 Review.
-
Biological Networks Underlying Abiotic Stress Tolerance in Temperate Crops--A Proteomic Perspective.Int J Mol Sci. 2015 Sep 1;16(9):20913-42. doi: 10.3390/ijms160920913. Int J Mol Sci. 2015. PMID: 26340626 Free PMC article. Review.
Cited by
-
Impacts, Tolerance, Adaptation, and Mitigation of Heat Stress on Wheat under Changing Climates.Int J Mol Sci. 2022 Mar 4;23(5):2838. doi: 10.3390/ijms23052838. Int J Mol Sci. 2022. PMID: 35269980 Free PMC article. Review.
-
Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects.Proteomes. 2022 May 22;10(2):17. doi: 10.3390/proteomes10020017. Proteomes. 2022. PMID: 35645375 Free PMC article. Review.
-
Comparative Proteomic Analysis of Grapevine Rootstock in Response to Waterlogging Stress.Front Plant Sci. 2021 Oct 29;12:749184. doi: 10.3389/fpls.2021.749184. eCollection 2021. Front Plant Sci. 2021. PMID: 34777428 Free PMC article.
-
Review: Proteomic Techniques for the Development of Flood-Tolerant Soybean.Int J Mol Sci. 2020 Oct 12;21(20):7497. doi: 10.3390/ijms21207497. Int J Mol Sci. 2020. PMID: 33053653 Free PMC article. Review.
-
Multi-Omics Uncover the Mechanism of Wheat under Heavy Metal Stress.Int J Mol Sci. 2022 Dec 15;23(24):15968. doi: 10.3390/ijms232415968. Int J Mol Sci. 2022. PMID: 36555610 Free PMC article. Review.
References
-
- Sayre KD, Ginkel MV, Rajaram S, Ortiz-Monasterio I. Annual Wheat Newsletter. 1994. Tolerance to waterlogging losses in spring bread wheat: effect of time of onset on expression; pp. 165–171.
-
- Samad A, Meisner CA, Saifuzzaman M, Ginekel MV. Waterlogging Tolerance. 2001.
-
- Collaku A, Harrison SA. Losses in wheat due to waterlogging. Crop Sci. 2002;42(2):444–450. doi: 10.2135/cropsci2002.4440. - DOI
-
- Trnka M, Rötter RP, Ruizramos M, Kersebaum KC, Olesen JE, Žalud Z, Semenov MA. Adverse weather conditions for European wheat production will become more frequent with climate change. Nat Clim Chang. 2014;4(7):637–643. doi: 10.1038/nclimate2242. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

