DHHC protein family targets different subsets of glioma stem cells in specific niches
- PMID: 30658672
- PMCID: PMC6339410
- DOI: 10.1186/s13046-019-1033-2
DHHC protein family targets different subsets of glioma stem cells in specific niches
Abstract
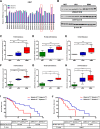
Background: Glioblastomas (GBM) comprise different subsets that exhibit marked heterogeneity and plasticity, leading to a lack of success of genomic profiling in guiding the development of precision medicine approaches against these tumors. Accordingly, there is an urgent need to investigate the regulatory mechanisms for different GBM subsets and identify novel biomarkers and therapeutic targets relevant in the context of GBM-specific niches. The DHHC family of proteins is associated tightly with the malignant development and progression of gliomas. However, the role of these proteins in the plasticity of GBM subsets remains unclear.
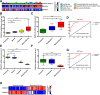
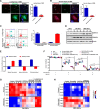
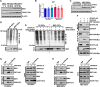
Methods: This study utilized human glioma proneural or mesenchymal stem cells as indicated. The effects of DHHC proteins on different GBM subsets were investigated through in vitro and in vivo assays (i.e., colony formation assay, flow cytometry assay, double immunofluorescence, western blot, and xenograft model). Western blot, co-immunoprecipitation, and liquid chromatograph mass spectrometer-mass spectrometry assays were used to detect the protein complexes of ZDHHC18 and ZDHHC23 in various GBM subtypes, and explore the mechanism of DHHC proteins in targeting different subsets of GSCs in specific niches.
Results: ZDHHC18 and ZDHHC23 could target the glioma stem cells of different GBM subsets in the context of their specific niches and regulate the cellular plasticity of these subtypes. Moreover, mechanistic investigations revealed that ZDHHC18 and ZDHHC23 competitively interact with a BMI1 E3 ligase, RNF144A, to regulate the polyubiquitination and accumulation of BMI1. These events contributed to the transition of glioma stem cells in GBM and cell survival under the stressful tumor microenvironment.
Conclusions: Our work highlights the role of DHHC proteins in the plasticity of GBM subsets and reveals that BMI1 represents a potential therapeutic target for human gliomas.
Keywords: BMI1; DHHC protein; Glioblastoma; Glioma stem cell.
Conflict of interest statement
Ethics approval and consent to participate
This study was reviewed and approved by the Institutional Review Board of the Animal Use and Care Committees at Hefei Institutes of Physical Science, CAS.
Consent for publication
All authors have agreed to publish this manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
A novel drug conjugate, NEO212, targeting proneural and mesenchymal subtypes of patient-derived glioma cancer stem cells.Cancer Lett. 2016 Feb 28;371(2):240-50. doi: 10.1016/j.canlet.2015.11.040. Epub 2015 Dec 9. Cancer Lett. 2016. PMID: 26683773
-
Gambogic acid impairs the maintenance and therapeutic resistance of glioma stem cells by targeting B-cell-specific Moloney leukemia virus insert site 1.Phytomedicine. 2024 Dec;135:156070. doi: 10.1016/j.phymed.2024.156070. Epub 2024 Sep 17. Phytomedicine. 2024. PMID: 39326139
-
miR128-1 inhibits the growth of glioblastoma multiforme and glioma stem-like cells via targeting BMI1 and E2F3.Oncotarget. 2016 Nov 29;7(48):78813-78826. doi: 10.18632/oncotarget.12385. Oncotarget. 2016. PMID: 27705931 Free PMC article.
-
Stem cell signature in glioblastoma: therapeutic development for a moving target.J Neurosurg. 2015 Feb;122(2):324-30. doi: 10.3171/2014.9.JNS132253. Epub 2014 Nov 14. J Neurosurg. 2015. PMID: 25397368 Review.
-
Extracellular vesicles in the biology of brain tumour stem cells--Implications for inter-cellular communication, therapy and biomarker development.Semin Cell Dev Biol. 2015 Apr;40:17-26. doi: 10.1016/j.semcdb.2015.02.011. Epub 2015 Feb 23. Semin Cell Dev Biol. 2015. PMID: 25721810 Review.
Cited by
-
MiR-146b-5p suppresses the malignancy of GSC/MSC fusion cells by targeting SMARCA5.Aging (Albany NY). 2020 Jul 6;12(13):13647-13667. doi: 10.18632/aging.103489. Epub 2020 Jul 6. Aging (Albany NY). 2020. PMID: 32632040 Free PMC article.
-
Novel peripheral blood diagnostic biomarkers screened by machine learning algorithms in ankylosing spondylitis.Front Genet. 2022 Nov 1;13:1032010. doi: 10.3389/fgene.2022.1032010. eCollection 2022. Front Genet. 2022. PMID: 36386830 Free PMC article.
-
Diverse Roles of Protein Palmitoylation in Cancer Progression, Immunity, Stemness, and Beyond.Cells. 2023 Sep 5;12(18):2209. doi: 10.3390/cells12182209. Cells. 2023. PMID: 37759431 Free PMC article. Review.
-
Clinical implication of cellular vaccine in glioma: current advances and future prospects.J Exp Clin Cancer Res. 2020 Nov 23;39(1):257. doi: 10.1186/s13046-020-01778-6. J Exp Clin Cancer Res. 2020. PMID: 33228738 Free PMC article. Review.
-
The role of E3 ubiquitin ligases in the development and progression of glioblastoma.Cell Death Differ. 2021 Feb;28(2):522-537. doi: 10.1038/s41418-020-00696-6. Epub 2021 Jan 11. Cell Death Differ. 2021. PMID: 33432111 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 81872066 and 31501171/National Natural Science Foundation of China
- 31571433 and 81773131/National Natural Science Foundation of China
- 1608085MH180/Natural Science Foundation of Anhui Province
- 1808085QH272/Natural Science Foundation of Anhui Province
- 2018CXFX004/the innovative program of Development Foundation of Hefei Center for Physical Science and Technology
LinkOut - more resources
Full Text Sources

