Host Range-Associated Clustering Based on Multilocus Variable-Number Tandem-Repeat Analysis, Phylotypes, and Virulence Genes of Atypical Enteropathogenic Escherichia coli Strains
- PMID: 30658974
- PMCID: PMC6414391
- DOI: 10.1128/AEM.02796-18
Host Range-Associated Clustering Based on Multilocus Variable-Number Tandem-Repeat Analysis, Phylotypes, and Virulence Genes of Atypical Enteropathogenic Escherichia coli Strains
Abstract
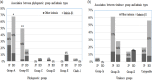
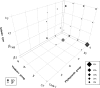
Atypical enteropathogenic Escherichia coli (aEPEC) strains (36 Japanese and 50 Bangladeshi) obtained from 649 poultry fecal samples were analyzed by molecular epidemiological methods. Clermont's phylogenetic typing showed that group A was more prevalent (58%, 50/86) than B1 (31%, 27/86). Intimin type β1, which is prevalent among human diarrheal patients, was predominant in both phylogroups B1 (81%, 22/27) and A (70%, 35/50). However, about 95% of B1-β1 strains belonged to virulence group I, and 77% of them were Japanese strains, while 17% (6/35) of A-β1 strains did. Multilocus variable-number tandem-repeat analysis (MLVA) distributed the strains into 52 distinct profiles, with Simpson's index of diversity (D) at 73%. When the data were combined with those of 142 previous strains from different sources, the minimum spanning tree formed five zones for porcine strains, poultry strains (excluding B1-β1), strains from healthy humans, bovine and human patient strains, and the B1-β1 poultry strains. Antimicrobial resistance to nalidixic acid was most common (74%) among the isolates. Sixty-eight percent of them demonstrated resistance to ≥3 antimicrobial agents, and most of them (91%) were from Bangladesh. The strains were assigned into two groups by hierarchical clustering. Correlation matrix analysis revealed that the virulence genes were negatively associated with antimicrobial resistance. The present study suggested that poultry, particularly Japanese poultry, could be another reservoir of aEPEC (phylogroup B1, virulence group I, and intimin type β1); however, poultry strains seem to be apart from patient strains that were closer to bovine strains. Bangladeshi aEPEC may be less virulent for humans but more resistant to antibiotics.IMPORTANCE Atypical enteropathogenic Escherichia coli (aEPEC) is a diarrheagenic type of E. coli, as it possesses the intimin gene (eae) for attachment and effacement on epithelium. Since aEPEC is ubiquitous even in developed countries, we previously used molecular epidemiological methods to discriminate aEPEC as a human pathogen. The present study assessed poultry as another source of human diarrheagenic aEPEC. Poultry could be the source of aEPEC (phylogroup B1, virulence group I, and intimin type β1) found among patient strains in Japan. However, the minimum spanning tree (MST) suggested that the strains from Japanese poultry were far from Japanese patient strains compared with the distance between bovine and patient strains. Bangladeshi avian strains seemed to be less diarrheagenic but are hazardous as a source of drug resistance genes.
Keywords: Escherichia coli; MLVA; drug resistance; molecular epidemiology; phylogeny; virulence.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114–2118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

