Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription
- PMID: 30659020
- PMCID: PMC8631248
- DOI: 10.1158/0008-5472.CAN-18-2505
Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription
Abstract
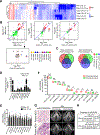
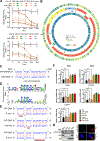
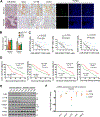
miRNAs that translocate from the nucleus to mitochondria are referred to as mitochondrial microRNAs (mitomiR). mitomiRs have been shown to modulate the translational activity of the mitochondrial genome, yet their role in mitochondrial DNA (mtDNA) transcription remains to be determined. Here we report that the mitomiR-2392 regulates chemoresistance in tongue squamous cell carcinoma (TSCC) cells by reprogramming metabolism via downregulation of oxidative phosphorylation and upregulation of glycolysis. These effects were mediated through partial inhibition of mtDNA transcription by mitomiR-2392 rather than through translational regulation. This repression required specific miRNA-mtDNA base pairing and Argonaute 2. mitomiR-2392 recognized target sequences in the H-strand and partially inhibited polycistronic mtDNA transcription in a cell-specific manner. A retrospective analysis of TSCC patient tumors revealed a significant association of miR-2392 and regulated mitochondrial gene expression with chemosensitivity and overall survival. The clinical relevance of targeted mitochondrial genes was consistently validated by The Cancer Genome Atlas RNA sequencing in multiple types of cancer. Our study revealed for the first time the role of mitomiR in mtDNA transcription and its contribution to the molecular basis of tumor cell metabolism and chemoresistance.Significance: These findings uncover a novel mechanism by which mitomiRNA regulates mitochondrial transcription and provide rationale for use of mitomiRNA and mtDNA-encoded genes to predict chemosensitivity and patient clinical prognosis.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures



Similar articles
-
Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation.Theranostics. 2019 Aug 12;9(20):5739-5754. doi: 10.7150/thno.37556. eCollection 2019. Theranostics. 2019. PMID: 31534516 Free PMC article.
-
Deregulation of the miR-222-ABCG2 regulatory module in tongue squamous cell carcinoma contributes to chemoresistance and enhanced migratory/invasive potential.Oncotarget. 2015 Dec 29;6(42):44538-50. doi: 10.18632/oncotarget.6253. Oncotarget. 2015. PMID: 26517090 Free PMC article.
-
Long Noncoding RNA MPRL Promotes Mitochondrial Fission and Cisplatin Chemosensitivity via Disruption of Pre-miRNA Processing.Clin Cancer Res. 2019 Jun 15;25(12):3673-3688. doi: 10.1158/1078-0432.CCR-18-2739. Epub 2019 Mar 18. Clin Cancer Res. 2019. PMID: 30885939 Free PMC article.
-
Mitochondria and cancer chemoresistance.Biochim Biophys Acta Bioenerg. 2017 Aug;1858(8):686-699. doi: 10.1016/j.bbabio.2017.01.012. Epub 2017 Feb 1. Biochim Biophys Acta Bioenerg. 2017. PMID: 28161329 Review.
-
Mitochondrial DNA and MitomiR Variations in Pancreatic Cancer: Potential Diagnostic and Prognostic Biomarkers.Int J Mol Sci. 2021 Sep 7;22(18):9692. doi: 10.3390/ijms22189692. Int J Mol Sci. 2021. PMID: 34575852 Free PMC article. Review.
Cited by
-
Zinc Finger Protein 521, Negatively Regulated by MicroRNA-204-5p, Promotes Proliferation, Motility and Invasion of Gastric Cancer Cells.Technol Cancer Res Treat. 2019 Jan 1;18:1533033819874783. doi: 10.1177/1533033819874783. Technol Cancer Res Treat. 2019. PMID: 31526099 Free PMC article.
-
Bta-miR-181d and Bta-miR-196a mediated proliferation, differentiation, and apoptosis in Bovine Myogenic Cells.J Anim Sci. 2024 Jan 3;102:skae142. doi: 10.1093/jas/skae142. J Anim Sci. 2024. PMID: 38766769 Free PMC article.
-
The Role of Mitochondria in the Chemoresistance of Pancreatic Cancer Cells.Cells. 2021 Feb 25;10(3):497. doi: 10.3390/cells10030497. Cells. 2021. PMID: 33669111 Free PMC article. Review.
-
Functional Role of Mitochondrial DNA in Cancer Progression.Int J Mol Sci. 2022 Jan 31;23(3):1659. doi: 10.3390/ijms23031659. Int J Mol Sci. 2022. PMID: 35163579 Free PMC article. Review.
-
MicroRNAs as Factors in Bidirectional Crosstalk Between Mitochondria and the Nucleus During Cellular Senescence.Front Physiol. 2021 Sep 9;12:734976. doi: 10.3389/fphys.2021.734976. eCollection 2021. Front Physiol. 2021. PMID: 34566699 Free PMC article. Review.
References
-
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. - PubMed
-
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. - PubMed
-
- Rodriguez-Enriquez S, Carreno-Fuentes L, Gallardo-Perez JC, Saavedra E, Quezada H, Vega A, et al. Oxidative phosphorylation is impaired by prolonged hypoxia in breast and possibly in cervix carcinoma. Int J Biochem Cell Biol. 2010;42:1744–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

