Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain
- PMID: 30659150
- PMCID: PMC6369821
- DOI: 10.1073/pnas.1811768116
Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain
Abstract
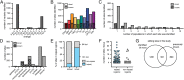
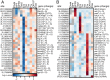
Adenosine-to-inosine (A-to-I) RNA editing, catalyzed by ADAR enzymes, is a ubiquitous mechanism that generates transcriptomic diversity. This process is particularly important for proper neuronal function; however, little is known about how RNA editing is dynamically regulated between the many functionally distinct neuronal populations of the brain. Here, we present a spatial RNA editing map in the Drosophila brain and show that different neuronal populations possess distinct RNA editing signatures. After purifying and sequencing RNA from genetically marked groups of neuronal nuclei, we identified a large number of editing sites and compared editing levels in hundreds of transcripts across nine functionally different neuronal populations. We found distinct editing repertoires for each population, including sites in repeat regions of the transcriptome and differential editing in highly conserved and likely functional regions of transcripts that encode essential neuronal genes. These changes are site-specific and not driven by changes in Adar expression, suggesting a complex, targeted regulation of editing levels in key transcripts. This fine-tuning of the transcriptome between different neurons by RNA editing may account for functional differences between distinct populations in the brain.
Keywords: Drosophila; RNA editing; neurons.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Palladino MJ, Keegan LP, O’Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

