Towards artificial intelligence in mental health by improving schizophrenia prediction with multiple brain parcellation ensemble-learning
- PMID: 30659193
- PMCID: PMC6386753
- DOI: 10.1038/s41537-018-0070-8
Towards artificial intelligence in mental health by improving schizophrenia prediction with multiple brain parcellation ensemble-learning
Abstract
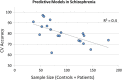
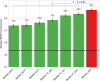
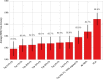
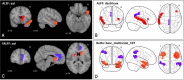
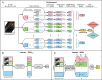
In the literature, there are substantial machine learning attempts to classify schizophrenia based on alterations in resting-state (RS) brain patterns using functional magnetic resonance imaging (fMRI). Most earlier studies modelled patients undergoing treatment, entailing confounding with drug effects on brain activity, and making them less applicable to real-world diagnosis at the point of first medical contact. Further, most studies with classification accuracies >80% are based on small sample datasets, which may be insufficient to capture the heterogeneity of schizophrenia, limiting generalization to unseen cases. In this study, we used RS fMRI data collected from a cohort of antipsychotic drug treatment-naive patients meeting DSM IV criteria for schizophrenia (N = 81) as well as age- and sex-matched healthy controls (N = 93). We present an ensemble model -- EMPaSchiz (read as 'Emphasis'; standing for 'Ensemble algorithm with Multiple Parcellations for Schizophrenia prediction') that stacks predictions from several 'single-source' models, each based on features of regional activity and functional connectivity, over a range of different a priori parcellation schemes. EMPaSchiz yielded a classification accuracy of 87% (vs. chance accuracy of 53%), which out-performs earlier machine learning models built for diagnosing schizophrenia using RS fMRI measures modelled on large samples (N > 100). To our knowledge, EMPaSchiz is first to be reported that has been trained and validated exclusively on data from drug-naive patients diagnosed with schizophrenia. The method relies on a single modality of MRI acquisition and can be readily scaled-up without needing to rebuild parcellation maps from incoming training images.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

