Arx Expression Suppresses Ventralization of the Developing Dorsal Forebrain
- PMID: 30659230
- PMCID: PMC6338776
- DOI: 10.1038/s41598-018-36194-6
Arx Expression Suppresses Ventralization of the Developing Dorsal Forebrain
Abstract
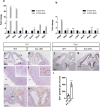
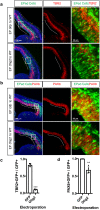
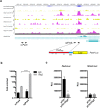
Early brain development requires a tight orchestration between neural tube patterning and growth. How pattern formation and brain growth are coordinated is incompletely understood. Previously we showed that aristaless-related homeobox (ARX), a paired-like transcription factor, regulates cortical progenitor pool expansion by repressing an inhibitor of cell cycle progression. Here we show that ARX participates in establishing dorsoventral identity in the mouse forebrain. In Arx mutant mice, ventral genes, including Olig2, are ectopically expressed dorsally. Furthermore, Gli1 is upregulated, suggesting an ectopic activation of SHH signaling. We show that the ectopic Olig2 expression can be repressed by blocking SHH signaling, implicating a role for SHH signaling in Olig2 induction. We further demonstrate that the ectopic Olig2 accounts for the reduced Pax6 and Tbr2 expression, both dorsal specific genes essential for cortical progenitor cell proliferation. These data suggest a link between the control of dorsoventral identity of progenitor cells and the control of their proliferation. In summary, our data demonstrate that ARX functions in a gene regulatory network integrating normal forebrain patterning and growth, providing important insight into how mutations in ARX can disrupt multiple aspects of brain development and thus generate a wide spectrum of neurodevelopmental phenotypes observed in human patients.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain.J Comp Neurol. 2005 Mar 14;483(3):292-303. doi: 10.1002/cne.20405. J Comp Neurol. 2005. PMID: 15682394
-
Arx together with FoxA2, regulates Shh floor plate expression.Dev Biol. 2014 Sep 1;393(1):137-48. doi: 10.1016/j.ydbio.2014.06.012. Epub 2014 Jun 23. Dev Biol. 2014. PMID: 24968361 Free PMC article.
-
Induction of oligodendrocyte progenitors in dorsal forebrain by intraventricular microinjection of FGF-2.Dev Biol. 2006 Sep 1;297(1):262-73. doi: 10.1016/j.ydbio.2006.05.017. Epub 2006 May 19. Dev Biol. 2006. PMID: 16782086
-
ARX: a gene for all seasons.Curr Opin Genet Dev. 2006 Jun;16(3):308-16. doi: 10.1016/j.gde.2006.04.003. Epub 2006 May 2. Curr Opin Genet Dev. 2006. PMID: 16650978 Review.
-
Further Delineation of Duplications of ARX Locus Detected in Male Patients with Varying Degrees of Intellectual Disability.Int J Mol Sci. 2022 Mar 13;23(6):3084. doi: 10.3390/ijms23063084. Int J Mol Sci. 2022. PMID: 35328505 Free PMC article. Review.
Cited by
-
Integrated Omic Analyses Identify Pathways and Transcriptomic Regulators Associated With Chemical Alterations of In Vitro Neural Network Formation.Toxicol Sci. 2022 Feb 28;186(1):118-133. doi: 10.1093/toxsci/kfab151. Toxicol Sci. 2022. PMID: 34927697 Free PMC article.
-
Specification of human brain regions with orthogonal gradients of WNT and SHH in organoids reveals patterning variations across cell lines.Cell Stem Cell. 2025 Jun 5;32(6):970-989.e11. doi: 10.1016/j.stem.2025.04.006. Epub 2025 May 1. Cell Stem Cell. 2025. PMID: 40315847
-
Development of a larval nervous system in the sea urchin.Curr Top Dev Biol. 2022;146:25-48. doi: 10.1016/bs.ctdb.2021.10.002. Epub 2021 Dec 3. Curr Top Dev Biol. 2022. PMID: 35152985 Free PMC article. Review.
-
Deregulation of microtubule organization and RNA metabolism in Arx models for lissencephaly and developmental epileptic encephalopathy.Hum Mol Genet. 2022 Jun 4;31(11):1884-1908. doi: 10.1093/hmg/ddac028. Hum Mol Genet. 2022. PMID: 35094084 Free PMC article.
-
Generation of FLAG-tagged Arx knock-in mouse model.Genesis. 2022 Jul;60(6-7):e23479. doi: 10.1002/dvg.23479. Epub 2022 Jun 3. Genesis. 2022. PMID: 35656878 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

