Classification of human Herpesviridae proteins using Domain-architecture Aware Inference of Orthologs (DAIO)
- PMID: 30660046
- PMCID: PMC6502252
- DOI: 10.1016/j.virol.2019.01.005
Classification of human Herpesviridae proteins using Domain-architecture Aware Inference of Orthologs (DAIO)
Abstract
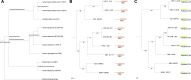
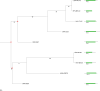
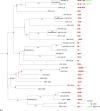
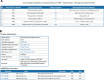
We developed a computational approach called Domain-architecture Aware Inference of Orthologs (DAIO) for the analysis of protein orthology by combining phylogenetic and protein domain-architecture information. Using DAIO, we performed a systematic study of the proteomes of all human Herpesviridae species to define Strict Ortholog Groups (SOGs). In addition to assessing the taxonomic distribution for each protein based on sequence similarity, we performed a protein domain-architecture analysis for every protein family and computationally inferred gene duplication events. While many herpesvirus proteins have evolved without any detectable gene duplications or domain rearrangements, numerous herpesvirus protein families do exhibit complex evolutionary histories. Some proteins acquired additional domains (e.g., DNA polymerase), whereas others show a combination of domain acquisition and gene duplication (e.g., betaherpesvirus US22 family), with possible functional implications. This novel classification system of SOGs for human Herpesviridae proteins is available through the Virus Pathogen Resource (ViPR, www.viprbrc.org).
Keywords: Comparative genomics; Domain architecture; Evolution; Gene duplication; Herpesviridae; Nomenclature; Ortholog; Phylogenetics; Protein domain; Protein family.
Copyright © 2019. Published by Elsevier Inc.
Figures


References
-
- Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye-AJee H., Cowley A., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Castro L.G., Figueira L., Garmiri P., Georghiou G., Gonzalez D., Hatton-Ellis E., Li W., Liu W., Lopez R., Luo J., Lussi Y., MacDougall A., Nightingale A., Palka B., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Speretta E., Turner E., Tyagi N., Volynkin V., Wardell T., Warner K., Watkins X., Zaru R., Zellner H., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., ArgoudPuy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. - DOI - PMC - PubMed
-
- Bridges K.G., Hua Q., Brigham-Burke M.R., Martin J.D., Hensley P., Dahl C.E., Digard P., Weiss M.A., Coen D.M. Secondary structure and structure-activity relationships of peptides corresponding to the subunit interface of herpes simplex virus DNA polymerase. J. Biol. Chem. 2000;275:472–478. doi: 10.1074/jbc.275.1.472. - DOI - PubMed

