Single-molecule FRET method to investigate the dynamics of transcription elongation through the nucleosome by RNA polymerase II
- PMID: 30660864
- PMCID: PMC6589119
- DOI: 10.1016/j.ymeth.2019.01.009
Single-molecule FRET method to investigate the dynamics of transcription elongation through the nucleosome by RNA polymerase II
Abstract
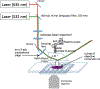
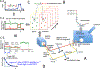
Transcription elongation through the nucleosome is a precisely coordinated activity to ensure timely production of RNA and accurate regulation of co-transcriptional histone modifications. Nucleosomes actively participate in transcription regulation at various levels and impose physical barriers to RNA polymerase II (RNAPII) during transcription elongation. Despite its high significance, the detailed dynamics of how RNAPII translocates along nucleosomal DNA during transcription elongation and how the nucleosome structure dynamically conforms to the changes necessary for RNAPII progression remain poorly understood. Transcription elongation through the nucleosome is a complex process and investigating the changes of the nucleosome structure during this process by ensemble measurements is daunting. This is because it is nearly impossible to synchronize elongation complexes within a nucleosome or a sub-nucleosome to a designated location at a high enough efficiency for desired sample homogeneity. Here we review our recently developed single-molecule FRET experimental system and method that has fulfilled this deficiency. With our method, one can follow the changes in the structure of individual nucleosomes during transcription elongation. We demonstrated that this method enables the detailed measurements of the kinetics of transcription elongation through the nucleosome and its regulation by a transcription factor, which can be easily extended to investigations of the roles of environmental variables and histone post-translational modifications in regulating transcription elongation.
Keywords: Nucleosome; RNA polymerase II; Single-molecule FRET; Transcription elongation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


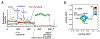
References
-
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD, Architecture of RNA polymerase II and implications for the transcription mechanism, Science (New York, N.Y.), 288 (2000) 640–649. - PubMed
-
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M, Nature of the nucleosomal barrier to RNA polymerase II, Mol Cell, 18 (2005) 97–108. - PubMed
-
- Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P, Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA, Mol Cell, 34 (2009) 710–721. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

