Co-Expression of a Chimeric Protease Inhibitor Secreted by a Tumor-Targeted Salmonella Protects Therapeutic Proteins from Proteolytic Degradation
- PMID: 30661346
- PMCID: PMC6883771
- DOI: 10.4014/jmb.1807.08036
Co-Expression of a Chimeric Protease Inhibitor Secreted by a Tumor-Targeted Salmonella Protects Therapeutic Proteins from Proteolytic Degradation
Abstract
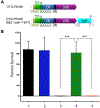
Sunflower trypsin inhibitor (SFTI) is a 14-amino-acid bicyclic peptide that contains a single internal disulfide bond. We initially constructed chimeras of SFTI with N-terminal secretion signals from the Escherichia coli OmpA and Pseudomonas aeruginosa ToxA, but only detected small amounts of protease inhibition resulting from these constructs. A substantially higher degree of protease inhibition was detected from a C-terminal SFTI fusion with E. coli YebF, which radiated more than a centimeter from an individual colony of E. coli using a culture-based inhibitor assay. Inhibitory activity was further improved in YebF-SFTI fusions by the addition of a trypsin cleavage signal immediately upstream of SFTI, and resulted in production of a 14-amino-acid, disulfide-bonded SFTI free in the culture supernatant. To assess the potential of the secreted SFTI to protect the ability of a cytotoxic protein to kill tumor cells, we utilized a tumor-selective form of the Pseudomonas ToxA (OTG-PE38K) alone and expressed as a polycistronic construct with YebF-SFTI in the tumor-targeted Salmonella VNP20009. When we assessed the ability of toxin-containing culture supernatants to kill MDA-MB-468 breast cancer cells, the untreated OTG-PE38K was able to eliminate all detectable tumor cells, while pretreatment with trypsin resulted in the complete loss of anticancer cytotoxicity. However, when OTG-PE38K was co-expressed with YebF-SFTI, cytotoxicity was completely retained in the presence of trypsin. These data demonstrate SFTI chimeras are secreted in a functional form and that co-expression of protease inhibitors with therapeutic proteins by tumor-targeted bacteria has the potential to enhance the activity of therapeutic proteins by suppressing their degradation within a proteolytic environment.
Keywords: OTG-PE38K; Protease inhibitors; VNP20009; YebF; sunflower trypsin inhibitor (SFTI); tumor-targeted Salmonella.
Conflict of interest statement
Conflict of Interest
DB has financial interest in Aviex Technologies and Magna Therapeutics, and receives royalties from Yale University. DB and DQ have interest in a patent application that includes subject materials from this report.
Figures




Similar articles
-
EGFR-targeted Chimeras of Pseudomonas ToxA released into the extracellular milieu by attenuated Salmonella selectively kill tumor cells.Biotechnol Bioeng. 2016 Dec;113(12):2698-2711. doi: 10.1002/bit.26026. Epub 2016 Jul 8. Biotechnol Bioeng. 2016. PMID: 27260220 Free PMC article.
-
Alanine scanning mutagenesis identifies surface amino acids on domain II of Pseudomonas exotoxin required for cytotoxicity, proper folding, and secretion into periplasm.J Biol Chem. 1992 Nov 15;267(32):23427-33. J Biol Chem. 1992. PMID: 1429683
-
Regulation of toxA and regA by the Escherichia coli fur gene and identification of a Fur homologue in Pseudomonas aeruginosa PA103 and PA01.Mol Microbiol. 1991 Nov;5(11):2823-31. doi: 10.1111/j.1365-2958.1991.tb01991.x. Mol Microbiol. 1991. PMID: 1779768
-
Pseudomonas exotoxin: chimeric toxins.J Biol Chem. 1989 Sep 15;264(26):15157-60. J Biol Chem. 1989. PMID: 2504717 Review.
-
Molecular Chimera in Cancer Drug Discovery: Beyond Antibody Therapy, Designing Grafted Stable Peptides Targeting Cancer.Int J Pept Res Ther. 2025;31(3):38. doi: 10.1007/s10989-025-10690-6. Epub 2025 Feb 17. Int J Pept Res Ther. 2025. PMID: 39974747 Free PMC article. Review.
Cited by
-
Tryptophanase Expressed by Salmonella Halts Breast Cancer Cell Growth In Vitro and Inhibits Production of Immunosuppressive Kynurenine.Microorganisms. 2023 May 22;11(5):1355. doi: 10.3390/microorganisms11051355. Microorganisms. 2023. PMID: 37317329 Free PMC article.
References
-
- Yoon J, Maruyama J, Kitamoto K. 2004. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl. Microbiol. Biotechnol 89: 747–759. - PubMed
-
- Scharpe S, De Meester I, Hendriks D, Vanhoof G, van Sande M, Vriend G. 1991. Proteases and their inhibitors: today and tomorrow. Biochimie 73: 121–126. - PubMed
-
- Jose J, Zangen D. 2005. Autodisplay of the protease inhibitor aprotinin in Escherichia coli. Biochem. Biophys. Res. Commun 333: 1218–1226. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

