The Oral Microbiome in the Elderly With Dental Caries and Health
- PMID: 30662876
- PMCID: PMC6328972
- DOI: 10.3389/fcimb.2018.00442
The Oral Microbiome in the Elderly With Dental Caries and Health
Abstract
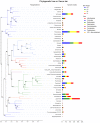
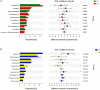
With the aging of the population, dental caries in the elderly has received increasing attention. A comprehensive study of the oral microbiome is required to understand its polymicrobial etiology. The results of previous studies are limited and remain controversial. In this study, subjects 60 years and older with and without caries were recruited. Unstimulated saliva and dental plaque were collected from each subject and the bacterial 16S rDNA was amplified using PCR and sequenced by Illumina MiSeq high-throughput sequencing. A total of 92 samples were collected from 24 caries patients and 22 healthy controls. Sequences clustered into 147,531 OTUs, representing 16 phyla, 29 classes, 49 orders, 79 families, 149 genera, and 305 species. All predominant phyla, including Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Actinobacteria, and Saccharibacteria, were largely consistent in different groups, but different relative abundances could be observed. The core microbiome was defined with 246 shared species among groups, which occupied 80.7% of all the species detected. Alpha diversity showed no significant differences in bacterial richness or diversity between caries patients and healthy controls, but distinction existed between samples collected from dental plaque and saliva. Beta diversity analysis was performed by PCoA and hierarchical clustering analysis, showing similar results that microorganisms vary between the two niches. The biomarkers of different groups were defined by LEfSe analysis to identify potential caries-related and health-related bacteria. The co-occurrence analysis of the predominant genera revealed significant interactions among oral microbiota and exhibited more complex and aggregated bacterial correlations in caries-free groups. Finally, the functional prediction of the microbiota present in oral samples was performed by PICRUSt, indicating vigorous microbial metabolism in the oral bacterial community. Our study provides thorough knowledge of the microbiological etiology of elderly individuals with caries and is expected to provide novel methods for its prevention and treatment.
Keywords: aged; biodiversity; dental caries; dental plaque; high-throughput nucleotide sequencing; microbiota; saliva.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

