Challenges in Stratifying the Molecular Variability of Patient-Derived Colon Tumor Xenografts
- PMID: 30662905
- PMCID: PMC6313970
- DOI: 10.1155/2018/2954208
Challenges in Stratifying the Molecular Variability of Patient-Derived Colon Tumor Xenografts
Abstract
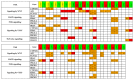
Colorectal cancer (CRC) is the second most common cancer in Europe and a leading cause of death worldwide. Patient-derived xenograft (PDX) models maintain complex intratumoral biology and heterogeneity and therefore remain the platform of choice for translational drug discovery. In this study, we implanted 37 primary CRC tumors and five CRC cell lines into NU/J mice to develop xenograft models. Primary tumors and established xenografts were histologically assessed and surveyed for genetic variants and gene expression using a panel of 409 cancer-related genes and RNA-seq, respectively. More than half of CRC tumors (20 out of 37, 54%) developed into a PDX. Histological assessment confirmed that PDX grading, stromal components, inflammation, and budding were consistent with those of the primary tumors. DNA sequencing identified an average of 0.14 variants per gene per sample. The percentage of mutated variants in PDXs increased with successive passages, indicating a decrease in clonal heterogeneity. Gene Ontology analyses of 4180 differentially expressed transcripts (adj. p value < 0.05) revealed overrepresentation of genes involved in cell division and catabolic processes among the transcripts upregulated in PDXs; downregulated transcripts were associated with GO terms related to extracellular matrix organization, immune responses, and angiogenesis. Neither a transcriptome-based consensus molecular subtype (CMS) classifier nor three other predictors reliably matched PDX molecular subtypes with those of the primary tumors. In sum, both genetic and transcriptomic profiles differed between donor tumors and PDXs, likely as a consequence of subclonal evolution at the early phase of xenograft development, making molecular stratification of PDXs challenging.
Figures




References
-
- Punt C. J. A., Koopman M., Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nature Reviews Clinical Oncology. 2017;14:235–246.235246 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

