Multiorgan segmentation using distance-aware adversarial networks
- PMID: 30662925
- PMCID: PMC6328005
- DOI: 10.1117/1.JMI.6.1.014001
Multiorgan segmentation using distance-aware adversarial networks
Abstract
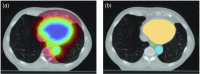
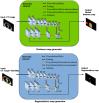
Segmentation of organs at risk (OAR) in computed tomography (CT) is of vital importance in radiotherapy treatment. This task is time consuming and for some organs, it is very challenging due to low-intensity contrast in CT. We propose a framework to perform the automatic segmentation of multiple OAR: esophagus, heart, trachea, and aorta. Different from previous works using deep learning techniques, we make use of global localization information, based on an original distance map that yields not only the localization of each organ, but also the spatial relationship between them. Instead of segmenting directly the organs, we first generate the localization map by minimizing a reconstruction error within an adversarial framework. This map that includes localization information of all organs is then used to guide the segmentation task in a fully convolutional setting. Experimental results show encouraging performance on CT scans of 60 patients totaling 11,084 slices in comparison with other state-of-the-art methods.
Keywords: convolutional neural networks; deep learning; distance map; generative adversarial networks; medical images; multiorgan; segmentation.
Figures








References
-
- Criminisi A., et al. , Regression Forests for Efficient Anatomy Detection and Localization in CT Studies, pp. 106–117, Springer, Berlin, Heidelberg: (2011).
-
- Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems 25, Pereira F., et al., Eds., Curran Associates, Inc., pp. 1097–1105 (2012).
-
- Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” CoRR arXiv:1409.1556 (2014).

