A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia
- PMID: 30664655
- PMCID: PMC6341102
- DOI: 10.1038/s41467-018-08147-0
A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia
Abstract
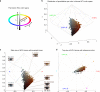
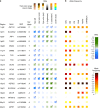
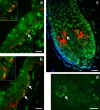
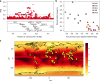
We report a genome-wide association scan in >6,000 Latin Americans for pigmentation of skin and eyes. We found eighteen signals of association at twelve genomic regions. These include one novel locus for skin pigmentation (in 10q26) and three novel loci for eye pigmentation (in 1q32, 20q13 and 22q12). We demonstrate the presence of multiple independent signals of association in the 11q14 and 15q13 regions (comprising the GRM5/TYR and HERC2/OCA2 genes, respectively) and several epistatic interactions among independently associated alleles. Strongest association with skin pigmentation at 19p13 was observed for an Y182H missense variant (common only in East Asians and Native Americans) in MFSD12, a gene recently associated with skin pigmentation in Africans. We show that the frequency of the derived allele at Y182H is significantly correlated with lower solar radiation intensity in East Asia and infer that MFSD12 was under selection in East Asians, probably after their split from Europeans.
Conflict of interest statement
J.C.C.-D. was employed by Living DNA from October 2017 to November 2018. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

