The Crohn's disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response
- PMID: 30666959
- PMCID: PMC6342529
- DOI: 10.7554/eLife.39982
The Crohn's disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response
Abstract
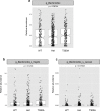
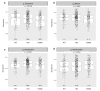
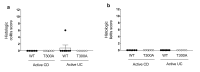
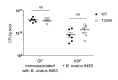
Inflammatory bowel disease (IBD) is driven by dysfunction between host genetics, the microbiota, and immune system. Knowledge gaps remain regarding how IBD genetic risk loci drive gut microbiota changes. The Crohn's disease risk allele ATG16L1 T300A results in abnormal Paneth cells due to decreased selective autophagy, increased cytokine release, and decreased intracellular bacterial clearance. To unravel the effects of ATG16L1 T300A on the microbiota and immune system, we employed a gnotobiotic model using human fecal transfers into ATG16L1 T300A knock-in mice. We observed increases in Bacteroides ovatus and Th1 and Th17 cells in ATG16L1 T300A mice. Association of altered Schaedler flora mice with B. ovatus specifically increased Th17 cells selectively in ATG16L1 T300A knock-in mice. Changes occur before disease onset, suggesting that ATG16L1 T300A contributes to dysbiosis and immune infiltration prior to disease symptoms. Our work provides insight for future studies on IBD subtypes, IBD patient treatment and diagnostics.
Keywords: ATG16L1T300A; Bacteroides; Bacteroides ovatus; Crohn's Disease; T cell; gut; immunology; infectious disease; inflammation; microbiology; mouse.
© 2019, Lavoie et al.
Conflict of interest statement
SL, KC, KL, HJ, HP, EC, MM, JL, CG, JD, JG, HV, AA, AK, RX No competing interests declared, WG Senior editor, eLife
Figures












References
-
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLOS Computational Biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. - DOI - PMC - PubMed
-
- Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, Shimizu N, Fujiyama Y. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. - DOI - PubMed
-
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host & Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. - DOI - PMC - PubMed
-
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK097485/NH/NIH HHS/United States
- DK105653/NH/NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- F31 DK105653/DK/NIDDK NIH HHS/United States
- R01 DK092405/DK/NIDDK NIH HHS/United States
- CA154426/NH/NIH HHS/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- DK110499/NH/NIH HHS/United States
- R01 CA154426/CA/NCI NIH HHS/United States
- Smith Family Foundation Award for Excellence in Biomedical Research/Richard and Susan Smith Family Foundation/International
- DK092405/NH/NIH HHS/United States
- R24 DK110499/DK/NIDDK NIH HHS/United States
- R01 DK097485/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

