Genome-wide analysis of genomic alterations induced by oxidative DNA damage in yeast
- PMID: 30668788
- PMCID: PMC6468167
- DOI: 10.1093/nar/gkz027
Genome-wide analysis of genomic alterations induced by oxidative DNA damage in yeast
Abstract
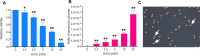
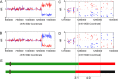
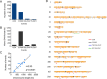
Oxidative DNA damage is a threat to genome stability. Using a genetic system in yeast that allows detection of mitotic recombination, we found that the frequency of crossovers is greatly elevated when cells are treated with hydrogen peroxide (H2O2). Using a combination of microarray analysis and genomic sequencing, we mapped the breakpoints of mitotic recombination events and other chromosome rearrangements at a resolution of about 1 kb. Gene conversions and crossovers were the two most common types of events, but we also observed deletions, duplications, and chromosome aneuploidy. In addition, H2O2-treated cells had elevated rates of point mutations (particularly A to T/T to A and C to G/G to C transversions) and small insertions/deletions (in/dels). In cells that underwent multiple rounds of H2O2 treatments, we identified a genetic alteration that resulted in improved H2O2 tolerance by amplification of the CTT1 gene that encodes cytosolic catalase T. Lastly, we showed that cells grown in the absence of oxygen have reduced levels of recombination. This study provided multiple novel insights into how oxidative stress affects genomic instability and phenotypic evolution in aerobic cells.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Hydrogen peroxide, a potent inducer of global genomic instability.Curr Genet. 2019 Aug;65(4):913-917. doi: 10.1007/s00294-019-00969-9. Epub 2019 Apr 8. Curr Genet. 2019. PMID: 30963245 Review.
-
Global Analysis of Furfural-Induced Genomic Instability Using a Yeast Model.Appl Environ Microbiol. 2019 Aug 29;85(18):e01237-19. doi: 10.1128/AEM.01237-19. Print 2019 Sep 15. Appl Environ Microbiol. 2019. PMID: 31300396 Free PMC article.
-
Aneuploidy drives genomic instability in yeast.Science. 2011 Aug 19;333(6045):1026-30. doi: 10.1126/science.1206412. Science. 2011. PMID: 21852501 Free PMC article.
-
High-resolution mapping of two types of spontaneous mitotic gene conversion events in Saccharomyces cerevisiae.Genetics. 2014 Sep;198(1):181-92. doi: 10.1534/genetics.114.167395. Epub 2014 Jul 1. Genetics. 2014. PMID: 24990991 Free PMC article.
-
Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae.DNA Repair (Amst). 2006 Sep 8;5(9-10):1010-20. doi: 10.1016/j.dnarep.2006.05.027. Epub 2006 Jun 23. DNA Repair (Amst). 2006. PMID: 16798113 Review.
Cited by
-
Hydrogen peroxide, a potent inducer of global genomic instability.Curr Genet. 2019 Aug;65(4):913-917. doi: 10.1007/s00294-019-00969-9. Epub 2019 Apr 8. Curr Genet. 2019. PMID: 30963245 Review.
-
Spontaneous and environment induced genomic alterations in yeast model.Cell Insight. 2024 Sep 26;4(1):100209. doi: 10.1016/j.cellin.2024.100209. eCollection 2025 Feb. Cell Insight. 2024. PMID: 39629481 Free PMC article. Review.
-
Genomic alterations of marine yeast Scheffersomyces spartinae under spontaneous and mutagenic conditions.BMC Genomics. 2025 Mar 25;26(1):297. doi: 10.1186/s12864-025-11479-z. BMC Genomics. 2025. PMID: 40133852 Free PMC article.
-
Functional interplay between the oxidative stress response and DNA damage checkpoint signaling for genome maintenance in aerobic organisms.J Microbiol. 2020 Feb;58(2):81-91. doi: 10.1007/s12275-020-9520-x. Epub 2019 Dec 23. J Microbiol. 2020. PMID: 31875928 Review.
-
Dynamics of DNA damage-induced nuclear inclusions are regulated by SUMOylation of Btn2.Nat Commun. 2024 Apr 13;15(1):3215. doi: 10.1038/s41467-024-47615-8. Nat Commun. 2024. PMID: 38615096 Free PMC article.
References
-
- Finkel T., Holbrook N.J.. Oxidants, oxidative stress and the biology of ageing. Nature. 2000; 408:239. - PubMed
-
- Herrero E., Ros J., Bellí G., Cabiscol E.. Redox control and oxidative stress in yeast cells. BBA-Gen. Subj. 2008; 1780:1217–1235. - PubMed
-
- Mizutani T., Sumida H., Sagawa Y., Okano Y., Masaki H.. ROS generation from the stratum corneum under UV irradiation. J. Dermatol. Sci. 2016; 84:e78–e79. - PubMed
-
- Fedoseeva I.V., Pyatrikas D.V., Stepanov A.V., Fedyaeva A.V., Varakina N.N., Rusaleva T.M., Borovskii G.B., Rikhvanov E.G.. The role of flavin-containing enzymes in mitochondrial membrane hyperpolarization and ROS production in respiring Saccharomyces cerevisiae cells under heat-shock conditions. Sci. Rep. 2017; 7:2586. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

