Marginal Likelihoods in Phylogenetics: A Review of Methods and Applications
- PMID: 30668834
- PMCID: PMC6701458
- DOI: 10.1093/sysbio/syz003
Marginal Likelihoods in Phylogenetics: A Review of Methods and Applications
Abstract
By providing a framework of accounting for the shared ancestry inherent to all life, phylogenetics is becoming the statistical foundation of biology. The importance of model choice continues to grow as phylogenetic models continue to increase in complexity to better capture micro- and macroevolutionary processes. In a Bayesian framework, the marginal likelihood is how data update our prior beliefs about models, which gives us an intuitive measure of comparing model fit that is grounded in probability theory. Given the rapid increase in the number and complexity of phylogenetic models, methods for approximating marginal likelihoods are increasingly important. Here, we try to provide an intuitive description of marginal likelihoods and why they are important in Bayesian model testing. We also categorize and review methods for estimating marginal likelihoods of phylogenetic models, highlighting several recent methods that provide well-behaved estimates. Furthermore, we review some empirical studies that demonstrate how marginal likelihoods can be used to learn about models of evolution from biological data. We discuss promising alternatives that can complement marginal likelihoods for Bayesian model choice, including posterior-predictive methods. Using simulations, we find one alternative method based on approximate-Bayesian computation to be biased. We conclude by discussing the challenges of Bayesian model choice and future directions that promise to improve the approximation of marginal likelihoods and Bayesian phylogenetics as a whole.
Keywords: Marginal likelihood; model choice; phylogenetics.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society of Systematic Biologists.
Figures
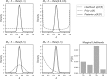

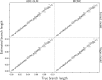
Similar articles
-
Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty.Syst Biol. 2016 Mar;65(2):250-64. doi: 10.1093/sysbio/syv083. Epub 2015 Nov 1. Syst Biol. 2016. PMID: 26526428 Free PMC article.
-
19 Dubious Ways to Compute the Marginal Likelihood of a Phylogenetic Tree Topology.Syst Biol. 2020 Mar 1;69(2):209-220. doi: 10.1093/sysbio/syz046. Syst Biol. 2020. PMID: 31504998 Free PMC article.
-
Assessing the Adequacy of Morphological Models Using Posterior Predictive Simulations.Syst Biol. 2025 Feb 10;74(1):34-52. doi: 10.1093/sysbio/syae055. Syst Biol. 2025. PMID: 39374100
-
Improving marginal likelihood estimation for Bayesian phylogenetic model selection.Syst Biol. 2011 Mar;60(2):150-60. doi: 10.1093/sysbio/syq085. Epub 2010 Dec 27. Syst Biol. 2011. PMID: 21187451 Free PMC article.
-
Systematic Exploration of the High Likelihood Set of Phylogenetic Tree Topologies.Syst Biol. 2020 Mar 1;69(2):280-293. doi: 10.1093/sysbio/syz047. Syst Biol. 2020. PMID: 31504997 Free PMC article.
Cited by
-
Evolutionary rate of SARS-CoV-2 increases during zoonotic infection of farmed mink.Virus Evol. 2023 Jan 10;9(1):vead002. doi: 10.1093/ve/vead002. eCollection 2023. Virus Evol. 2023. PMID: 36751428 Free PMC article.
-
Optimizing representations for integrative structural modeling using Bayesian model selection.Bioinformatics. 2024 Mar 4;40(3):btae106. doi: 10.1093/bioinformatics/btae106. Bioinformatics. 2024. PMID: 38391029 Free PMC article.
-
Estimating effective population size changes from preferentially sampled genetic sequences.PLoS Comput Biol. 2020 Oct 12;16(10):e1007774. doi: 10.1371/journal.pcbi.1007774. eCollection 2020 Oct. PLoS Comput Biol. 2020. PMID: 33044955 Free PMC article.
-
The comparative biogeography of Philippine geckos challenges predictions from a paradigm of climate-driven vicariant diversification across an island archipelago.Evolution. 2019 Jun;73(6):1151-1167. doi: 10.1111/evo.13754. Epub 2019 May 9. Evolution. 2019. PMID: 31017301 Free PMC article.
-
Under pressure: phenotypic divergence and convergence associated with microhabitat adaptations in Triatominae.Parasit Vectors. 2021 Apr 8;14(1):195. doi: 10.1186/s13071-021-04647-z. Parasit Vectors. 2021. PMID: 33832518 Free PMC article.
References
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19:716–723.
-
- Arima S., Tardella L.. 2012. Improved harmonic mean estimator for phylogenetic model evidence. J. Comput. Biol. 19:418–438. - PubMed
-
- Arima S., Tardella L.. 2014. Inflated density ratio (IDR) method for estimating marginal likelihoods in Bayesian phylogenetics. In: Chen M.-H., Kuo L., Lewis P.O., editors. Bayesian phylogenetics: methods, algorithms, and applications, Chapter 3 Boca Raton (FL): CRC Press; p. 25–57.
-
- Baele G., Lemey P.. 2013. Bayesian evolutionary model testing in the phylogenomics era: matching model complexity with computational efficiency. Bioinformatics. 29:1970–1979. - PubMed
-
- Baele G., Lemey P.. 2014. Bayesian model selection in phylogenetics and genealogy-based population genetics. In: Chen M.-H., Kuo L., Lewis P.O., editors. Bayesian phylogenetics: methods, algorithms, and applications, Chapter 4. Boca Raton (FL): CRC Press; p. 59–93.

