DNA methylation GrimAge strongly predicts lifespan and healthspan
- PMID: 30669119
- PMCID: PMC6366976
- DOI: 10.18632/aging.101684
DNA methylation GrimAge strongly predicts lifespan and healthspan
Abstract
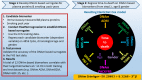
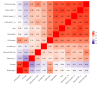
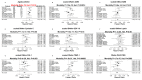
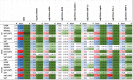
It was unknown whether plasma protein levels can be estimated based on DNA methylation (DNAm) levels, and if so, how the resulting surrogates can be consolidated into a powerful predictor of lifespan. We present here, seven DNAm-based estimators of plasma proteins including those of plasminogen activator inhibitor 1 (PAI-1) and growth differentiation factor 15. The resulting predictor of lifespan, DNAm GrimAge (in units of years), is a composite biomarker based on the seven DNAm surrogates and a DNAm-based estimator of smoking pack-years. Adjusting DNAm GrimAge for chronological age generated novel measure of epigenetic age acceleration, AgeAccelGrim.Using large scale validation data from thousands of individuals, we demonstrate that DNAm GrimAge stands out among existing epigenetic clocks in terms of its predictive ability for time-to-death (Cox regression P=2.0E-75), time-to-coronary heart disease (Cox P=6.2E-24), time-to-cancer (P= 1.3E-12), its strong relationship with computed tomography data for fatty liver/excess visceral fat, and age-at-menopause (P=1.6E-12). AgeAccelGrim is strongly associated with a host of age-related conditions including comorbidity count (P=3.45E-17). Similarly, age-adjusted DNAm PAI-1 levels are associated with lifespan (P=5.4E-28), comorbidity count (P= 7.3E-56) and type 2 diabetes (P=2.0E-26). These DNAm-based biomarkers show the expected relationship with lifestyle factors including healthy diet and educational attainment.Overall, these epigenetic biomarkers are expected to find many applications including human anti-aging studies.
Keywords: DNA methylation; epigenetics; mortality; proteomics.
Conflict of interest statement
Figures



References
-
- Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S, Wilson JG, Reiner AP, Maierhofer A, et al.. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY). 2018; 10:1758–75. 10.18632/aging.101508 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201300048C/HL/NHLBI NIH HHS/United States
- HHSN268201300049C/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201600001C/HL/NHLBI NIH HHS/United States
- HHSN268201300046C/HL/NHLBI NIH HHS/United States
- U54 GM115428/GM/NIGMS NIH HHS/United States
- U01 AG060908/AG/NIA NIH HHS/United States
- HHSN268201600002C/HL/NHLBI NIH HHS/United States
- R01 HL116446/HL/NHLBI NIH HHS/United States
- R01 ES020836/ES/NIEHS NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- HHSN268201600018C/HL/NHLBI NIH HHS/United States
- HHSN268201600003C/HL/NHLBI NIH HHS/United States
- R01 AG029451/AG/NIA NIH HHS/United States
- HHSN268201300047C/HL/NHLBI NIH HHS/United States
- HHSN268201300050C/HL/NHLBI NIH HHS/United States
- HHSN268201600004C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

