NUP214 in Leukemia: It's More than Transport
- PMID: 30669574
- PMCID: PMC6356203
- DOI: 10.3390/cells8010076
NUP214 in Leukemia: It's More than Transport
Abstract
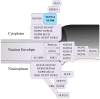
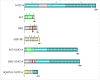
NUP214 is a component of the nuclear pore complex (NPC) with a key role in protein and mRNA nuclear export. Chromosomal translocations involving the NUP214 locus are recurrent in acute leukemia and frequently fuse the C-terminal region of NUP214 with SET and DEK, two chromatin remodeling proteins with roles in transcription regulation. SET-NUP214 and DEK-NUP214 fusion proteins disrupt protein nuclear export by inhibition of the nuclear export receptor CRM1, which results in the aberrant accumulation of CRM1 protein cargoes in the nucleus. SET-NUP214 is primarily associated with acute lymphoblastic leukemia (ALL), whereas DEK-NUP214 exclusively results in acute myeloid leukemia (AML), indicating different leukemogenic driver mechanisms. Secondary mutations in leukemic blasts may contribute to the different leukemia outcomes. Additional layers of complexity arise from the respective functions of SET and DEK in transcription regulation and chromatin remodeling, which may drive malignant hematopoietic transformation more towards ALL or AML. Another, less frequent fusion protein involving the C terminus of NUP214 results in the sequestosome-1 (SQSTM1)-NUP214 chimera, which was detected in ALL. SQSTM1 is a ubiquitin-binding protein required for proper autophagy induction, linking the NUP214 fusion protein to yet another cellular mechanism. The scope of this review is to summarize the general features of NUP214-related leukemia and discuss how distinct chromosomal translocation partners can influence the cellular effects of NUP214 fusion proteins in leukemia.
Keywords: CRM1; DEK; HOX genes; NUP214; SET; chromatin remodeling; leukemia; nucleocytoplasmic transport; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ferlay J.S.I., Ervik M., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. [(accessed on 24 November 2018)]; Available online: http://globocan.iarc.fr.
-
- The National Cancer Institute. [(accessed on 24 November 2018)]; Available online: www.cancer.gov.
-
- Cancer Research, UK. [(accessed on 24 November 2018)]; Available online: http://www.cancerresearchuk.org.
-
- Leukemia & Lymphoma Society. [(accessed on 5 November 2018)]; Available online: http://www.lls.org/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

