Roles of Autophagy-Related Genes in the Pathogenesis of Inflammatory Bowel Disease
- PMID: 30669622
- PMCID: PMC6356351
- DOI: 10.3390/cells8010077
Roles of Autophagy-Related Genes in the Pathogenesis of Inflammatory Bowel Disease
Abstract
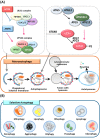
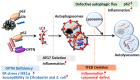
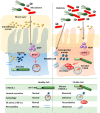
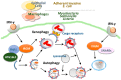
Autophagy is an intracellular catabolic process that is essential for a variety of cellular responses. Due to its role in the maintenance of biological homeostasis in conditions of stress, dysregulation or disruption of autophagy may be linked to human diseases such as inflammatory bowel disease (IBD). IBD is a complicated inflammatory colitis disorder; Crohn's disease and ulcerative colitis are the principal types. Genetic studies have shown the clinical relevance of several autophagy-related genes (ATGs) in the pathogenesis of IBD. Additionally, recent studies using conditional knockout mice have led to a comprehensive understanding of ATGs that affect intestinal inflammation, Paneth cell abnormality and enteric pathogenic infection during colitis. In this review, we discuss the various ATGs involved in macroautophagy and selective autophagy, including ATG16L1, IRGM, LRRK2, ATG7, p62, optineurin and TFEB in the maintenance of intestinal homeostasis. Although advances have been made regarding the involvement of ATGs in maintaining intestinal homeostasis, determining the precise contribution of autophagy has remained elusive. Recent efforts based on direct targeting of ATGs and autophagy will further facilitate the development of new therapeutic opportunities for IBD.
Keywords: ATGs; autophagy; inflammatory bowel diseases; intestinal homeostasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

