Double-Stranded RNA High-Throughput Sequencing Reveals a New Cytorhabdovirus in a Bean Golden Mosaic Virus-Resistant Common Bean Transgenic Line
- PMID: 30669683
- PMCID: PMC6357046
- DOI: 10.3390/v11010090
Double-Stranded RNA High-Throughput Sequencing Reveals a New Cytorhabdovirus in a Bean Golden Mosaic Virus-Resistant Common Bean Transgenic Line
Abstract
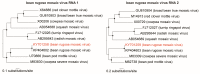
Using double-strand RNA (dsRNA) high-throughput sequencing, we identified five RNA viruses in a bean golden mosaic virus (BGMV)-resistant common bean transgenic line with symptoms of viral infection. Four of the identified viruses had already been described as infecting common bean (cowpea mild mottle virus, bean rugose mosaic virus, Phaseolus vulgaris alphaendornavirus 1, and Phaseolus vulgaris alphaendornavirus 2) and one is a putative new plant rhabdovirus (genus Cytorhabdovirus), tentatively named bean-associated cytorhabdovirus (BaCV). The BaCV genome presented all five open reading frames (ORFs) found in most rhabdoviruses: nucleoprotein (N) (ORF1) (451 amino acids, aa), phosphoprotein (P) (ORF2) (445 aa), matrix (M) (ORF4) (287 aa), glycoprotein (G) (ORF5) (520 aa), and an RNA-dependent RNA polymerase (L) (ORF6) (114 aa), as well as a putative movement protein (P3) (ORF3) (189 aa) and the hypothetical small protein P4. The predicted BaCV proteins were compared to homologous proteins from the closest cytorhabdoviruses, and a low level of sequence identity (15⁻39%) was observed. The phylogenetic analysis shows that BaCV clustered with yerba mate chlorosis-associated virus (YmCaV) and rice stripe mosaic virus (RSMV). Overall, our results provide strong evidence that BaCV is indeed a new virus species in the genus Cytorhabdovirus (family Rhabdoviridae), the first rhabdovirus to be identified infecting common bean.
Keywords: Phaseolus vulgaris; common bean; cytorhabdovirus; dsRNA high throughput sequencing; rhabdovirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Comment in
-
Letter to the Editor: Bean-Associated Cytorhabdovirus and Papaya Cytorhabdovirus are Strains of the Same Virus.Viruses. 2019 Mar 7;11(3):230. doi: 10.3390/v11030230. Viruses. 2019. PMID: 30866430 Free PMC article.
References
-
- FAOSTAT Food and Agriculture Organization of the United Nations-Statistics Division. [(accessed on 14 December 2018)];2018 Available online: http://www.fao.org/faostat/en/#home.
-
- Faria J.C., Aragão F.J.L., Souza T.L.P.O., Quintela E.D., Kitajima E.W., Ribeiro S.G. Golden Mosaic of Common Beans in Brazil: Management with a Transgenic Approach. APS Features. 2016 doi: 10.1094/APSFeature-2016-10. - DOI
-
- Okada R., Yong C.K., Valverde R.A., Sabanadzovic S., Aoki N., Hotate S., Kiyota E., Moriyama H., Fukuhara T. Molecular characterization of two evolutionarily distinct endornaviruses co-infecting common bean (Phaseolus vulgaris) J. Gen. Virol. 2013;94:220–229. doi: 10.1099/vir.0.044487-0. - DOI - PubMed
-
- Gilbertson R.L., Faria J.C., Ahlquist P., Maxwell D.P. Genetic Diversity in Geminiviruses Causing Bean Golden Mosaic Disease: The Nucleotide-Sequence of the Infectious Cloned DNA Components of a Brazilian Isolate of Bean Golden Mosaic Geminivirus. Phytopathology. 1993;83:709–715. doi: 10.1094/Phyto-83-709. - DOI
-
- Faria J.C., Gilbertson R.L., Hanson S.F., Morales F.J., Ahlquist P., Loniello A.O., Maxwell D.P. Bean golden mosaic geminivirus type II isolates from the Dominican Republic and Guatemala: Nucleotide sequences, infectious pseudorecombinants and phylogenetic relationships. Phytopathology. 1994;84:321–329. doi: 10.1094/Phyto-84-321. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

