Cul4 ubiquitin ligase cofactor DCAF12 promotes neurotransmitter release and homeostatic plasticity
- PMID: 30670470
- PMCID: PMC6400570
- DOI: 10.1083/jcb.201805099
Cul4 ubiquitin ligase cofactor DCAF12 promotes neurotransmitter release and homeostatic plasticity
Abstract
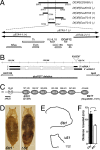
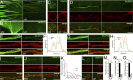
We genetically characterized the synaptic role of the Drosophila homologue of human DCAF12, a putative cofactor of Cullin4 (Cul4) ubiquitin ligase complexes. Deletion of Drosophila DCAF12 impairs larval locomotion and arrests development. At larval neuromuscular junctions (NMJs), DCAF12 is expressed presynaptically in synaptic boutons, axons, and nuclei of motor neurons. Postsynaptically, DCAF12 is expressed in muscle nuclei and facilitates Cul4-dependent ubiquitination. Genetic experiments identified several mechanistically independent functions of DCAF12 at larval NMJs. First, presynaptic DCAF12 promotes evoked neurotransmitter release. Second, postsynaptic DCAF12 negatively controls the synaptic levels of the glutamate receptor subunits GluRIIA, GluRIIC, and GluRIID. The down-regulation of synaptic GluRIIA subunits by nuclear DCAF12 requires Cul4. Third, presynaptic DCAF12 is required for the expression of synaptic homeostatic potentiation. We suggest that DCAF12 and Cul4 are critical for normal synaptic function and plasticity at larval NMJs.
© 2019 Patrón et al.
Figures







References
-
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., and Zheng N.. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 443:590–593. - PubMed
-
- Brand A.H., and Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

