Advances in understanding DNA processing and protection at stalled replication forks
- PMID: 30670471
- PMCID: PMC6446843
- DOI: 10.1083/jcb.201809012
Advances in understanding DNA processing and protection at stalled replication forks
Abstract
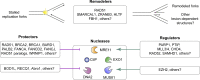
The replisome, the molecular machine dedicated to copying DNA, encounters a variety of obstacles during S phase. Without a proper response to this replication stress, the genome becomes unstable, leading to disease, including cancer. The immediate response is localized to the stalled replisome and includes protection of the nascent DNA. A number of recent studies have provided insight into the factors recruited to and responsible for protecting stalled replication forks. In response to replication stress, the SNF2 family of DNA translocases has emerged as being responsible for remodeling replication forks in vivo. The protection of stalled replication forks requires the cooperation of RAD51, BRCA1, BRCA2, and many other DNA damage response proteins. In the absence of these fork protection factors, fork remodeling renders them vulnerable to degradation by nucleases and helicases, ultimately compromising genome integrity. In this review, we focus on the recent progress in understanding the protection, processing, and remodeling of stalled replication forks in mammalian cells.
© 2019 Rickman and Smogorzewska.
Figures




References
-
- Ahuja A.K., Jodkowska K., Teloni F., Bizard A.H., Zellweger R., Herrador R., Ortega S., Hickson I.D., Altmeyer M., Mendez J., and Lopes M.. 2016. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7:10660 10.1038/ncomms10660 - DOI - PMC - PubMed
-
- Bacquin A., Pouvelle C., Siaud N., Perderiset M., Salomé-Desnoulez S., Tellier-Lebegue C., Lopez B., Charbonnier J.B., and Kannouche P.L.. 2013. The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 41:6501–6513. 10.1093/nar/gkt397 - DOI - PMC - PubMed
-
- Barazas M., Annunziato S., Pettitt S.J., de Krijger I., Ghezraoui H., Roobol S.J., Lutz C., Frankum J., Song F.F., Brough R., et al. . 2018. The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Reports. 23:2107–2118. 10.1016/j.celrep.2018.04.046 - DOI - PMC - PubMed
-
- Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., et al. . 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20:347–354. 10.1038/nsmb.2501 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

