Methylation Biomarker Panel Performance in EsophaCap Cytology Samples for Diagnosing Barrett's Esophagus: A Prospective Validation Study
- PMID: 30670490
- PMCID: PMC6594757
- DOI: 10.1158/1078-0432.CCR-18-3696
Methylation Biomarker Panel Performance in EsophaCap Cytology Samples for Diagnosing Barrett's Esophagus: A Prospective Validation Study
Abstract
Purpose: Barrett's esophagus is the only known precursor of esophageal adenocarcinoma (EAC). Although endoscopy and biopsy are standard methods for Barrett's esophagus diagnosis, their high cost and risk limit their use as a screening modality. Here, we sought to develop a Barrett's esophagus detection method based on methylation status in cytology samples captured by EsophaCap using a streamlined sensitive technique, methylation on beads (MOB).
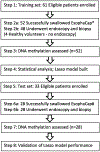
Experimental design: We conducted a prospective cohort study on 80 patients (52 in the training set; 28 in the test set). We used MOB to extract and bisulfite-convert DNA, followed by quantitative methylation-specific PCR to assess methylation levels of 8 previously selected candidate markers. Lasso regression was applied to establish a prediction model in the training set, which was then tested on the independent test set.
Results: In the training set, five of eight candidate methylation biomarkers (p16, HPP1, NELL1, TAC1, and AKAP12) were significantly higher in Barrett's esophagus patients than in controls. We built a four-biomarker-plus-age lasso regression model for Barrett's esophagus diagnosis. The AUC was 0.894, with sensitivity 94.4% [95% confidence interval (CI), 71%-99%] and specificity 62.2% (95% CI, 44.6%-77.3%) in the training set. This model also performed with high accuracy for Barrett's esophagus diagnosis in an independent test set: AUC = 0.929 (P < 0.001; 95% CI, 0.810%-1%), with sensitivity=78.6% (95% CI, 48.8%-94.3%) and specificity = 92.8% (95% CI, 64.1%-99.6%).
Conclusions: EsophaCap, in combination with an epigenetic biomarker panel and the MOB method, is a promising, well-tolerated, low-cost esophageal sampling strategy for Barrett's esophagus diagnosis. This approach merits further prospective studies in larger populations.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures

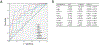


Comment in
-
Novel Barrett's esophagus screening assays based on swallowable devices: will they change the game?Transl Gastroenterol Hepatol. 2019 Apr 19;4:25. doi: 10.21037/tgh.2019.04.01. eCollection 2019. Transl Gastroenterol Hepatol. 2019. PMID: 31143846 Free PMC article. No abstract available.
-
To BE or not to BE: non-invasive screening for Barrett's esophagus, dysplasia and adenocarcinoma.Transl Gastroenterol Hepatol. 2019 May 15;4:31. doi: 10.21037/tgh.2019.04.08. eCollection 2019. Transl Gastroenterol Hepatol. 2019. PMID: 31231698 Free PMC article. No abstract available.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. - PubMed
-
- Spechler SJ. Barrett esophagus and risk of esophageal cancer: a clinical review. JAMA. 2013;310:627–36. - PubMed
-
- El-Serag HB, Naik AD, Duan Z, Shakhatreh M, Helm A, Pathak A, et al. Surveillance endoscopy is associated with improved outcomes of oesophageal adenocarcinoma detected in patients with Barrett’s oesophagus. Gut. 2015. - PubMed
-
- Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. The New England journal of medicine. 2011;365:1375–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

