Atypical Hemolytic Listeria innocua Isolates Are Virulent, albeit Less than Listeria monocytogenes
- PMID: 30670551
- PMCID: PMC6434133
- DOI: 10.1128/IAI.00758-18
Atypical Hemolytic Listeria innocua Isolates Are Virulent, albeit Less than Listeria monocytogenes
Abstract
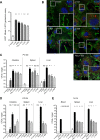
Listeria innocua is considered a nonpathogenic Listeria species. Natural atypical hemolytic L. innocua isolates have been reported but have not been characterized in detail. Here, we report the genomic and functional characterization of representative isolates from the two known natural hemolytic L. innocua clades. Whole-genome sequencing confirmed the presence of Listeria pathogenicity islands (LIPI) characteristic of Listeria monocytogenes species. Functional assays showed that LIPI-1 and inlA genes are transcribed, and the corresponding gene products are expressed and functional. Using in vitro and in vivo assays, we show that atypical hemolytic L. innocua is virulent, can actively cross the intestinal epithelium, and spreads systemically to the liver and spleen, albeit to a lesser degree than the reference L. monocytogenes EGDe strain. Although human exposure to hemolytic L. innocua is likely rare, these findings are important for food safety and public health. The presence of virulence traits in some L. innocua clades supports the existence of a common virulent ancestor of L. monocytogenes and L. innocua.
Keywords: L. innocua; LIPI; inlA; listeriosis; virulence.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- Charlier C, Perrodeau É, Leclercq A, Cazenave B, Pilmis B, Henry B, Lopes A, Maury MM, Moura A, Goffinet F, Dieye HB, Thouvenot P, Ungeheuer MN, Tourdjman M, Goulet V, de Valk H, Lortholary O, Ravaud P, Lecuit M, MONALISA Study Group. 2017. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 17:510–519. doi:10.1016/S1473-3099(16)30521-7. - DOI - PubMed
-
- Favaro M, Sarmati L, Sancesario G, Fontana C. 2014. First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep 1:1–5. doi:10.1099/jmmcr.0.003103. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

