Transcriptional repressor REST drives lineage stage-specific chromatin compaction at Ptch1 and increases AKT activation in a mouse model of medulloblastoma
- PMID: 30670636
- PMCID: PMC8939892
- DOI: 10.1126/scisignal.aan8680
Transcriptional repressor REST drives lineage stage-specific chromatin compaction at Ptch1 and increases AKT activation in a mouse model of medulloblastoma
Abstract
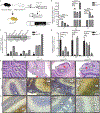
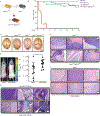
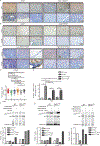
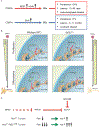
In medulloblastomas (MBs), the expression and activity of RE1-silencing transcription factor (REST) is increased in tumors driven by the sonic hedgehog (SHH) pathway, specifically the SHH-α (children 3 to 16 years) and SHH-β (infants) subgroups. Neuronal maturation is greater in SHH-β than SHH-α tumors, but both correlate with poor overall patient survival. We studied the contribution of REST to MB using a transgenic mouse model (RESTTG ) wherein conditional NeuroD2-controlled REST transgene expression in lineage-committed Ptch1 +/- cerebellar granule neuron progenitors (CGNPs) accelerated tumorigenesis and increased penetrance and infiltrative disease. This model revealed a neuronal maturation context-specific antagonistic interplay between the transcriptional repressor REST and the activator GLI1 at Ptch1 Expression of Arrb1, which encodes β-arrestin1 (a GLI1 inhibitor), was substantially reduced in proliferating and, to a lesser extent, lineage-committed RESTTG cells compared with wild-type proliferating CGNPs. Lineage-committed RESTTG cells also had decreased GLI1 activity and increased histone H3K9 methylation at the Ptch1 locus, which correlated with premature silencing of Ptch1 These cells also had decreased expression of Pten, which encodes a negative regulator of the kinase AKT. Expression of PTCH1 and GLI1 were less, and ARRB1 was somewhat greater, in patient SHH-β than SHH-α MBs, whereas that of PTEN was similarly lower in both subtypes than in others. Inhibition of histone modifiers or AKT reduced proliferation and induced apoptosis, respectively, in cultured REST-high MB cells. Our findings linking REST to differentiation-specific chromatin remodeling, PTCH1 silencing, and AKT activation in MB tissues reveal potential subgroup-specific therapeutic targets for MB patients.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures




References
-
- Coluccia D, Figuereido C, Isik S, Smith C, Rutka JT, Medulloblastoma: Tumor Biology and Relevance to Treatment and Prognosis Paradigm. Current neurology and neuroscience reports 16, 43 (2016). - PubMed
-
- Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM, Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123, 473–484 (2012). - PMC - PubMed
-
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM, Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123, 465–472 (2012). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

