Microbe-focused glycan array screening platform
- PMID: 30670663
- PMCID: PMC6369816
- DOI: 10.1073/pnas.1800853116
Microbe-focused glycan array screening platform
Abstract
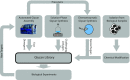
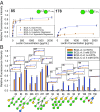
Interactions between glycans and glycan binding proteins are essential for numerous processes in all kingdoms of life. Glycan microarrays are an excellent tool to examine protein-glycan interactions. Here, we present a microbe-focused glycan microarray platform based on oligosaccharides obtained by chemical synthesis. Glycans were generated by combining different carbohydrate synthesis approaches including automated glycan assembly, solution-phase synthesis, and chemoenzymatic methods. The current library of more than 300 glycans is as diverse as the mammalian glycan array from the Consortium for Functional Glycomics and, due to its microbial focus, highly complementary. This glycan platform is essential for the characterization of various classes of glycan binding proteins. Applications of this glycan array platform are highlighted by the characterization of innate immune receptors and bacterial virulence factors as well as the analysis of human humoral immunity to pathogenic glycans.
Keywords: antiglycan antibodies; bacterial lectins; glycan arrays; immune receptors; microbial antigens.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

